Meiosis (; from Ancient Greek μείωσις 'lessening', is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one copy of each chromosome. Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a zygote, a cell with two copies of each chromosome again.

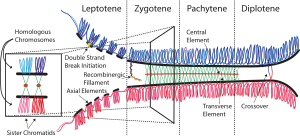
Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.
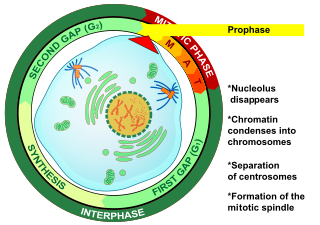
Prophase is the first stage of cell division in both mitosis and meiosis. Beginning after interphase, DNA has already been replicated when the cell enters prophase. The main occurrences in prophase are the condensation of the chromatin reticulum and the disappearance of the nucleolus.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) interchromosomal recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes ; & (2) intrachromosomal recombination, occurring through crossing over.

A pair of homologous chromosomes, or homologs, is a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci, where they provide points along each chromosome that enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance, which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.

A heteroduplex is a double-stranded (duplex) molecule of nucleic acid originated through the genetic recombination of single complementary strands derived from different sources, such as from different homologous chromosomes or even from different organisms.

Synapsis or Syzygy is the pairing of two chromosomes that occurs during meiosis. It allows matching-up of homologous pairs prior to their segregation, and possible chromosomal crossover between them. Synapsis takes place during prophase I of meiosis. When homologous chromosomes synapse, their ends are first attached to the nuclear envelope. These end-membrane complexes then migrate, assisted by the extranuclear cytoskeleton, until matching ends have been paired. Then the intervening regions of the chromosome are brought together, and may be connected by a protein-DNA complex called the synaptonemal complex. During synapsis, autosomes are held together by the synaptonemal complex along their whole length, whereas for sex chromosomes, this only takes place at one end of each chromosome.

A chromomere, also known as an idiomere, is one of the serially aligned beads or granules of a eukaryotic chromosome, resulting from local coiling of a continuous DNA thread. Chromomeres are regions of chromatin that have been compacted through localized contraction. In areas of chromatin with the absence of transcription, condensing of DNA and protein complexes will result in the formation of chromomeres. It is visible on a chromosome during the prophase of meiosis and mitosis. Giant banded (Polytene) chromosomes resulting from the replication of the chromosomes and the synapsis of homologs without cell division is a process called endomitosis. These chromosomes consist of more than 1000 copies of the same chromatid that are aligned and produce alternating dark and light bands when stained. The dark bands are the chromomere.
Zygotene is the second stage of prophase I during meiosis, the specialized cell division that reduces the chromosome number by half to produce haploid gametes. It follows the Leptotene stage and is followed by Pachytene stage.
The pachytene stage, also known as pachynema, is the third stage of prophase I during meiosis, the specialized cell division that reduces chromosome number by half to produce haploid gametes. It follows the zygotene stage and is followed by the stage Diplotene

In cellular biology, a bivalent is one pair of chromosomes in a tetrad. A tetrad is the association of a pair of homologous chromosomes physically held together by at least one DNA crossover. This physical attachment allows for alignment and segregation of the homologous chromosomes in the first meiotic division. In most organisms, each replicated chromosome elicits formation of DNA double-strand breaks during the leptotene phase. These breaks are repaired by homologous recombination, that uses the homologous chromosome as a template for repair. The search for the homologous target, helped by numerous proteins collectively referred as the synaptonemal complex, cause the two homologs to pair, between the leptotene and the pachytene phases of meiosis I.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

MutS protein homolog 4 is a protein that in humans is encoded by the MSH4 gene.

HORMA domain-containing protein 1 (HORMAD1) also known as cancer/testis antigen 46 (CT46) is a protein that in humans is encoded by the HORMAD1 gene.
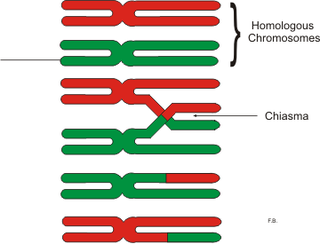
In genetics, a chiasma is the point of contact, the physical link, between two (non-sister) chromatids belonging to homologous chromosomes. At a given chiasma, an exchange of genetic material can occur between both chromatids, what is called a chromosomal crossover, but this is much more frequent during meiosis than mitosis. In meiosis, absence of a chiasma generally results in improper chromosomal segregation and aneuploidy.

The meiotic recombination checkpoint monitors meiotic recombination during meiosis, and blocks the entry into metaphase I if recombination is not efficiently processed.
The leptotene stage, also known as leptonema, is the first of five substages of prophase I during meiosis, the specialized cell division that reduces the chromosome number by half to produce haploid gametes in sexually reproducing organisms.

Structural maintenance of chromosomes protein 1B (SMC-1B) is a protein that in humans is encoded by the SMC1B gene. SMC proteins engage in chromosome organization and can be broken into 3 groups based on function which are cohesins, condensins, and DNA repair. SMC-1B belongs to a family of proteins required for chromatid cohesion and DNA recombination during meiosis and mitosis. SMC1B protein appears to participate with other cohesins REC8, STAG3 and SMC3 in sister-chromatid cohesion throughout the whole meiotic process in human oocytes.

Stromal antigen 3 is a protein that in humans is encoded by the STAG3 gene. STAG3 protein is a component of a cohesin complex that regulates the separation of sister chromatids specifically during meiosis. STAG3 appears to be paramount in sister-chromatid cohesion throughout the meiotic process in human oocytes and spermatocytes.
Abby F. Dernburg is a professor of Cell and Developmental Biology at the University of California, Berkeley, an Investigator of the Howard Hughes Medical Institute, and a Faculty Senior Scientist at Lawrence Berkeley National Laboratory.