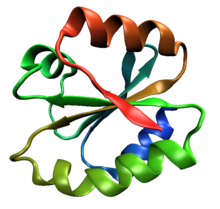
The beta sheet is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies.

Cysteine is a semiessential proteinogenic amino acid with the formula HOOC−CH(−NH2)−CH2−SH. The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. Cysteine is chiral, but both D and L-cysteine are found in nature. L‑Cysteine is a protein monomer in all biota, and D-cysteine acts as a signaling molecule in mammalian nervous systems. Cysteine is named after its discovery in urine, which comes from the urinary bladder or cyst, from Greek κύστη kýsti, "bladder".
In chemistry, a disulfide is a compound containing a R−S−S−R′ functional group or the S2−
2 anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups.

In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
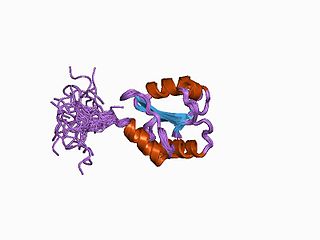
Protein disulfide isomerase, or PDI, is an enzyme in the endoplasmic reticulum (ER) in eukaryotes and the periplasm of bacteria that catalyzes the formation and breakage of disulfide bonds between cysteine residues within proteins as they fold. This allows proteins to quickly find the correct arrangement of disulfide bonds in their fully folded state, and therefore the enzyme acts to catalyze protein folding.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
Thioredoxin reductases are enzymes that reduce thioredoxin (Trx). Two classes of thioredoxin reductase have been identified: one class in bacteria and some eukaryotes and one in animals. In bacteria TrxR also catalyzes the reduction of glutaredoxin like proteins known as NrdH. Both classes are flavoproteins which function as homodimers. Each monomer contains a FAD prosthetic group, a NADPH binding domain, and an active site containing a redox-active disulfide bond.

Thioredoxin is a class of small redox proteins known to be present in all organisms. It plays a role in many important biological processes, including redox signaling. In humans, thioredoxins are encoded by TXN and TXN2 genes. Loss-of-function mutation of either of the two human thioredoxin genes is lethal at the four-cell stage of the developing embryo. Although not entirely understood, thioredoxin is linked to medicine through their response to reactive oxygen species (ROS). In plants, thioredoxins regulate a spectrum of critical functions, ranging from photosynthesis to growth, flowering and the development and germination of seeds. Thioredoxins play a role in cell-to-cell communication.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.
Oxidative protein folding is a process that is responsible for the formation of disulfide bonds between cysteine residues in proteins. The driving force behind this process is a redox reaction, in which electrons pass between several proteins and finally to a terminal electron acceptor.

ER oxidoreductin 1 (Ero1) is an oxidoreductase enzyme that catalyses the formation and isomerization of protein disulfide bonds in the endoplasmic reticulum (ER) of eukaryotes. ER Oxidoreductin 1 (Ero1) is a conserved, luminal, glycoprotein that is tightly associated with the ER membrane, and is essential for the oxidation of protein dithiols. Since disulfide bond formation is an oxidative process, the major pathway of its catalysis has evolved to utilise oxidoreductases, which become reduced during the thiol-disulfide exchange reactions that oxidise the cysteine thiol groups of nascent polypeptides. Ero1 is required for the introduction of oxidising equivalents into the ER and their direct transfer to protein disulfide isomerase (PDI), thereby ensuring the correct folding and assembly of proteins that contain disulfide bonds in their native state.
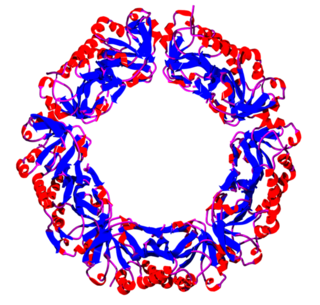
Peroxiredoxins are a ubiquitous family of antioxidant enzymes that also control cytokine-induced peroxide levels and thereby mediate signal transduction in mammalian cells. The family members in humans are PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6. The physiological importance of peroxiredoxins is indicated by their relative abundance. Their function is the reduction of peroxides, specifically hydrogen peroxide, alkyl hydroperoxides, and peroxynitrite.

Protein disulfide-isomerase A3 (PDIA3), also known as glucose-regulated protein, 58-kD (GRP58), is an isomerase enzyme encoded by the autosomal gene PDIA3 in humans. This protein localizes to the endoplasmic reticulum (ER) and interacts with lectin chaperones calreticulin and calnexin (CNX) to modulate folding of newly synthesized glycoproteins. It is thought that complexes of lectins and this protein mediate protein folding by promoting formation of disulfide bonds in their glycoprotein substrates.
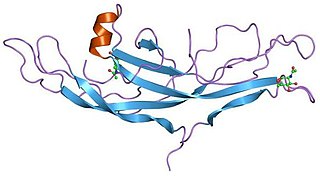
A cystine knot is a protein structural motif containing three disulfide bridges. The sections of polypeptide that occur between two of them form a loop through which a third disulfide bond passes, forming a rotaxane substructure. The cystine knot motif stabilizes protein structure and is conserved in proteins across various species. There are three types of cystine knot, which differ in the topology of the disulfide bonds:

DsbA is a bacterial thiol disulfide oxidoreductase (TDOR). DsbA is a key component of the Dsb family of enzymes. DsbA catalyzes intrachain disulfide bond formation as peptides emerge into the cell's periplasm.
Thioredoxins are small disulfide-containing redox proteins that have been found in all the kingdoms of living organisms. Thioredoxin serves as a general protein disulfide oxidoreductase. It interacts with a broad range of proteins by a redox mechanism based on reversible oxidation of 2 cysteine thiol groups to a disulfide, accompanied by the transfer of 2 electrons and 2 protons. The net result is the covalent interconversion of a disulfide and a dithiol.

DsbC (Disulfide bond C) is a prokaryotic disulfide bond isomerase. The formation of native disulfide bonds play an important role in the proper folding of proteins and stabilize tertiary structures of the protein. DsbC is one of 6 proteins in the Dsb family in prokaryotes. The other proteins are DsbA, DsbB, DsbD, DsbE and DsbG. These enzymes work in tandem with each other to form disulfide bonds during the expression of proteins. DsbC and DsbG act as proofreaders of the disulfide bonds that are formed. They break non-native disulfide bonds that were formed and act as chaperones for the formation of native disulfide bonds. The isomerization of disulfide bonds occurs in the periplasm.

Ferredoxin-thioredoxin reductase EC 1.8.7.2, systematic name ferredoxin:thioredoxin disulfide oxidoreductase, is a [4Fe-4S] protein that plays an important role in the ferredoxin/thioredoxin regulatory chain. It catalyzes the following reaction:
Huwentoxins (HWTX) are a group of neurotoxic peptides found in the venom of the Chinese bird spider Haplopelma schmidti. The species was formerly known as Haplopelma huwenum, Ornithoctonus huwena and Selenocosmia huwena. While structural similarity can be found among several of these toxins, HWTX as a group possess high functional diversity.

Galactose oxidase is an enzyme that catalyzes the oxidation of D-galactose in some species of fungi.