Defective interfering particles (DIPs), also known as defective interfering viruses, are spontaneously generated virus mutants in which a critical portion of the particle's genome has been lost due to defective replication or non-homologous recombination. The mechanism of their formation is presumed to be as a result of template-switching during replication of the viral genome, although non-replicative mechanisms involving direct ligation of genomic RNA fragments have also been proposed. DIPs are derived from and associated with their parent virus, and particles are classed as DIPs if they are rendered non-infectious due to at least one essential gene of the virus being lost or severely damaged as a result of the defection. A DIP can usually still penetrate host cells, but requires another fully functional virus particle to co-infect a cell with it, in order to provide the lost factors.

Reoviridae is a family of double-stranded RNA viruses. Member viruses have a wide host range, including vertebrates, invertebrates, plants, protists and fungi. They lack lipid envelopes and package their segmented genome within multi-layered capsids. Lack of a lipid envelope has allowed three-dimensional structures of these large complex viruses to be obtained, revealing a structural and likely evolutionary relationship to the cystovirus family of bacteriophage. There are currently 97 species in this family, divided among 15 genera in two subfamilies. Reoviruses can affect the gastrointestinal system and respiratory tract. The name "reo-" is an acronym for "respiratory enteric orphan" viruses. The term "orphan virus" refers to the fact that some of these viruses have been observed not associated with any known disease. Even though viruses in the family Reoviridae have more recently been identified with various diseases, the original name is still used.
Cauliflower mosaic virus (CaMV) is a member of the genus Caulimovirus, one of the six genera in the family Caulimoviridae, which are pararetroviruses that infect plants. Pararetroviruses replicate through reverse transcription just like retroviruses, but the viral particles contain DNA instead of RNA.

Rubella virus (RuV) is the pathogenic agent of the disease rubella, transmitted only between humans via the respiratory route, and is the main cause of congenital rubella syndrome when infection occurs during the first weeks of pregnancy.
A satellite is a subviral agent that depends on the coinfection of a host cell with a helper virus for its replication. Satellites can be divided into two major classes: satellite viruses and satellite nucleic acids. Satellite viruses, which are most commonly associated with plants, are also found in mammals, arthropods, and bacteria. They encode structural proteins to enclose their genetic material, which are therefore distinct from the structural proteins of their helper viruses. Satellite nucleic acids, in contrast, do not encode their own structural proteins, but instead are encapsulated by proteins encoded by their helper viruses. The genomes of satellites range upward from 359 nucleotides in length for satellite tobacco ringspot virus RNA (STobRV).

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.

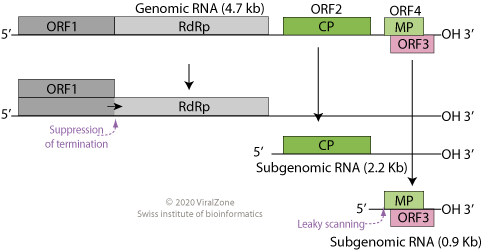
Tombusviridae is a family of single-stranded positive sense RNA plant viruses. There are three subfamilies, 17 genera, and 95 species in this family. The name is derived from Tomato bushy stunt virus (TBSV).
Tombusvirus is a genus of viruses, in the family Tombusviridae. Plants serve as natural hosts. There are 17 species in this genus. Symptoms associated with this genus include mosaic. The name of the genus comes from Tomato bushy stunt virus.

Agroinfiltration is a method used in plant biology and especially lately in plant biotechnology to induce transient expression of genes in a plant, or isolated leaves from a plant, or even in cultures of plant cells, in order to produce a desired protein. In the method, a suspension of Agrobacterium tumefaciens is introduced into a plant leaf by direct injection or by vacuum infiltration, or brought into association with plant cells immobilised on a porous support, whereafter the bacteria transfer the desired gene into the plant cells via transfer of T-DNA. The main benefit of agroinfiltration when compared to the more traditional plant transformation is speed and convenience, although yields of the recombinant protein are generally also higher and more consistent.
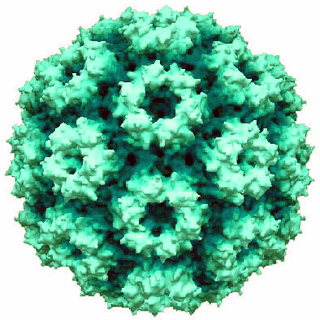
Cowpea chlorotic mottle virus, known by the abbreviation CCMV, is a virus that specifically infects the cowpea plant, or black-eyed pea. The leaves of infected plants develop yellow spots, hence the name "chlorotic". Similar to its "brother" virus, Cowpea mosaic virus (CPMV), CCMV is produced in high yield in plants. In the natural host, viral particles can be produced at 1–2 mg per gram of infected leaf tissue. Belonging to the bromovirus genus, cowpea chlorotic mottle virus (CCMV) is a small spherical plant virus. Other members of this genus include the brome mosaic virus (BMV) and the broad bean mottle virus (BBMV).

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.
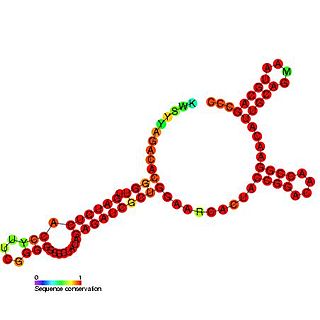
Tombusvirus 3′ UTR is an important cis-regulatory region of the Tombus virus genome.
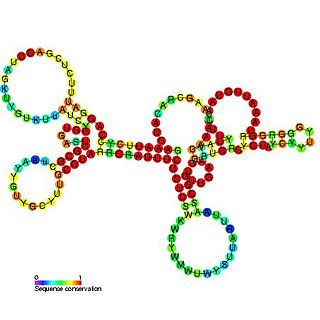
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.
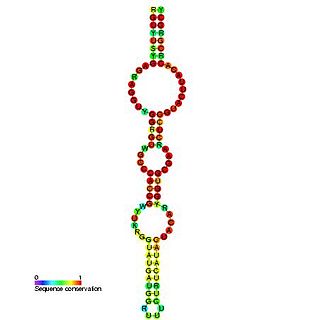
In virology, the tombusvirus internal replication element (IRE) is a segment of RNA located within the region coding for p92 polymerase. This element is essential for viral replication; specifically, it is thought to be required at an early stage of replication, such as template recruitment and/or replicase complex assembly.
Rice hoja blanca tenuivirus (RHBV), meaning "white leaf rice virus", is a plant virus in the family Phenuiviridae. RHBV causes Hoja blanca disease (HBD), which affects the leaves of the rice plant Oryza sativa, stunting the growth of the plant or killing it altogether. RHBV is carried by an insect vector, Tagosodes orizicolus, a type of planthopper. The virus is found in South America, Mexico, throughout Central America, the Caribbean region, and the southern United States. In South America, the disease is endemic to Colombia, Venezuela, Ecuador, Peru, Suriname, French Guiana and Guyana.
Potato mop-top virus (PMTV) is a plant pathogenic virus transmitted through the vector Spongospora subterranea that affects potatoes. PMTV belongs to family of Virgaviridae, and the genus Pomovirus. The virus was first identified in 1966 by Calvert and Harrison in Britain, and is now reported in many other potato cultivating regions of the world including U.S.A., Canada, China, Pakistan, Japan, South American countries and many parts of Europe. Many disease management systems have been found to be ineffective against the virus, although a combination of sanitation and vector controls seems to work well.
Turnip crinkle virus (TCV) is a plant pathogenic virus of the family Tombusviridae. It was first isolated from turnip.
Sweet potato feathery mottle virus (SPFMV) is a member of the genus Potyvirus in the family Potyviridae. It is most widely recognized as one of the most regularly occurring causal agents of sweet potato viral disease (SPVD) and is currently observed in every continent except Antarctica. The number of locations where it is found is still increasing; generally, it is assumed that the virus is present wherever its host is. The virus has four strains that are found in varying parts of the world.
Idaeovirus is a genus of positive-sense ssRNA viruses that contains two species: Raspberry bushy dwarf virus (RBDV) and Privet idaeovirus. RBDV has two host-dependent clades: one for raspberries; the other for grapevines. Infections are a significant agricultural burden, resulting in decreased yield and quality of crops. RBDV has a synergistic relation with Raspberry leaf mottle virus, with co-infection greatly amplifying the concentration of virions in infected plants. The virus is transmitted via pollination with RBDV-infected pollen grains that first infect the stigma before causing systemic infection.

RNA silencing suppressor p19 is a protein expressed from the ORF4 gene in the genome of tombusviruses. These viruses are positive-sense single-stranded RNA viruses that infect plant cells, in which RNA silencing forms a widespread and robust antiviral defense system. The p19 protein serves as a counter-defense strategy, specifically binding the 19- to 21-nucleotide double-stranded RNAs that function as small interfering RNA (siRNA) in the RNA silencing system. By sequestering siRNA, p19 suppresses RNA silencing and promotes viral proliferation. The p19 protein is considered a significant virulence factor and a component of an evolutionary arms race between plants and their pathogens.