
A haplotype is a group of alleles in an organism that are inherited together from a single parent, and a haplogroup is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism mutation. More specifically, a haplotype is a combination of alleles at different chromosomal regions that are closely linked and that tend to be inherited together. As a haplogroup consists of similar haplotypes, it is usually possible to predict a haplogroup from haplotypes. Haplogroups pertain to a single line of descent. As such, membership of a haplogroup, by any individual, relies on a relatively small proportion of the genetic material possessed by that individual.

Genetics and archaeogenetics of South Asia is the study of the genetics and archaeogenetics of the ethnic groups of South Asia. It aims at uncovering these groups' genetic histories. The geographic position of the Indian subcontinent makes its biodiversity important for the study of the early dispersal of anatomically modern humans across Asia.

In human genetics, Haplogroup J-M172 or J2 is a Y-chromosome haplogroup which is a subclade (branch) of haplogroup J-M304. Haplogroup J-M172 is common in modern populations in Western Asia, Central Asia, South Asia, Southern Europe, Northwestern Iran and North Africa. It is thought that J-M172 may have originated in the Caucasus, Anatolia and/or Western Iran.
Haplogroup T is a human mitochondrial DNA (mtDNA) haplogroup. It is believed to have originated around 25,100 years ago in the Near East.

Haplogroup R is a widely distributed human mitochondrial DNA (mtDNA) haplogroup. Haplogroup R is associated with the peopling of Eurasia after about 70,000 years ago, and is distributed in modern populations throughout the world outside of sub-Saharan Africa.
Haplogroup I is a human mitochondrial DNA (mtDNA) haplogroup. It is believed to have originated about 21,000 years ago, during the Last Glacial Maximum (LGM) period in West Asia. The haplogroup is unusual in that it is now widely distributed geographically, but is common in only a few small areas of East Africa, West Asia and Europe. It is especially common among the El Molo and Rendille peoples of Kenya, various regions of Iran, the Lemko people of Slovakia, Poland and Ukraine, the island of Krk in Croatia, the department of Finistère in France and some parts of Scotland and Ireland.

Haplogroup L-M20 is a human Y-DNA haplogroup, which is defined by SNPs M11, M20, M61 and M185. As a secondary descendant of haplogroup K and a primary branch of haplogroup LT, haplogroup L currently has the alternative phylogenetic name of K1a, and is a sibling of haplogroup T.

In human genetics, a human mitochondrial DNA haplogroup is a haplogroup defined by differences in human mitochondrial DNA. Haplogroups are used to represent the major branch points on the mitochondrial phylogenetic tree. Understanding the evolutionary path of the female lineage has helped population geneticists trace the matrilineal inheritance of modern humans back to human origins in Africa and the subsequent spread around the globe.
Haplogroup L0 is a human mitochondrial DNA (mtDNA) haplogroup.

In human mitochondrial genetics, Haplogroup Y is a human mitochondrial DNA (mtDNA) haplogroup.
In human mitochondrial genetics, Haplogroup G is a human mitochondrial DNA (mtDNA) haplogroup.
In the context of the recent African origin of modern humans, the Southern Dispersal scenario refers to the early migration along the southern coast of Asia, from the Arabian Peninsula via Persia and India to Southeast Asia and Oceania. Alternative names include the "southern coastal route" or "rapid coastal settlement", with later descendants of those migrations eventually colonizing the rest of Eastern Eurasia, the remainder of Oceania, and the Americas.
In human mitochondrial genetics, Haplogroup M30 is a human mitochondrial DNA (mtDNA) haplogroup.
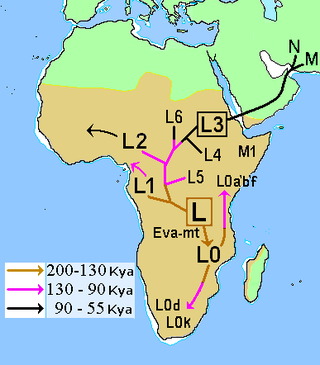
In human mitochondrial genetics, L is the mitochondrial DNA macro-haplogroup that is at the root of the anatomically modern human mtDNA phylogenetic tree. As such, it represents the most ancestral mitochondrial lineage of all currently living modern humans, also dubbed "Mitochondrial Eve".

The peopling of India refers to the migration of Homo sapiens into the Indian subcontinent. Anatomically modern humans settled India in multiple waves of early migrations, over tens of millennia. The first migrants came with the Coastal Migration/Southern Dispersal 65,000 years ago, whereafter complex migrations within South and Southeast Asia took place. West Asian (Iranian) hunter-gatherers migrated to South Asia after the Last Glacial Period but before the onset of farming. Together with ancient South Asian hunter-gatherers they formed the population of the Indus Valley Civilisation (IVC).

Genetic studies on the Sinhalese is part of population genetics investigating the origins of the Sinhalese population.
Y-DNA haplogroups in populations of South Asia are haplogroups of the male Y-chromosome found in South Asian populations.
In human mitochondrial genetics, haplogroup M18 is a human mitochondrial DNA (mtDNA) haplogroup. It is an India-specific lineage.
The study of the genetics and archaeogenetics of the Gujarati people of India aims at uncovering these people's genetic history. According to the 1000 Genomes Project, "Gujarati" is a general term used to describe people who trace their ancestry to the region of Gujarat, located in the northwestern part of the Indian subcontinent, and who speak the Gujarati language, an Indo-European language. They have some genetic commonalities as well as differences with other ethnic groups of India.









