Polyomaviridae is a family of viruses whose natural hosts are primarily mammals and birds. As of 2020, there are six recognized genera and 117 species, five of which are unassigned to a genus. 14 species are known to infect humans, while others, such as Simian Virus 40, have been identified in humans to a lesser extent. Most of these viruses are very common and typically asymptomatic in most human populations studied. BK virus is associated with nephropathy in renal transplant and non-renal solid organ transplant patients, JC virus with progressive multifocal leukoencephalopathy, and Merkel cell virus with Merkel cell cancer.
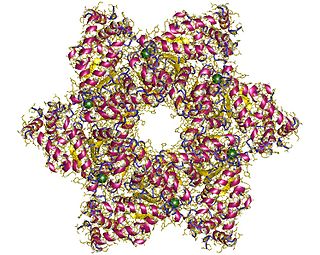
SV40 large T antigen is a hexamer protein that is a dominant-acting oncoprotein derived from the polyomavirus SV40. TAg is capable of inducing malignant transformation of a variety of cell types. The transforming activity of TAg is due in large part to its perturbation of the retinoblastoma (pRb) and p53 tumor suppressor proteins. In addition, TAg binds to several other cellular factors, including the transcriptional co-activators p300 and CBP, which may contribute to its transformation function. Similar proteins from related viruses are known as large tumor antigen in general.
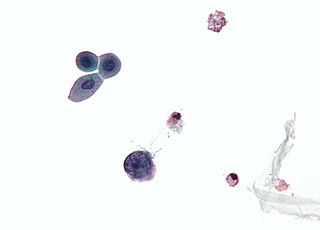
Merkel cell polyomavirus was first described in January 2008 in Pittsburgh, Pennsylvania. It was the first example of a human viral pathogen discovered using unbiased metagenomic next-generation sequencing with a technique called digital transcriptome subtraction. MCV is one of seven currently known human oncoviruses. It is suspected to cause the majority of cases of Merkel cell carcinoma, a rare but aggressive form of skin cancer. Approximately 80% of Merkel cell carcinoma (MCC) tumors have been found to be infected with MCV. MCV appears to be a common—if not universal—infection of older children and adults. It is found in respiratory secretions suggesting that it may be transmitted by a respiratory route. But it also can be found shedding from healthy skin, and in gastrointestinal tract tissues and elsewhere, and so its precise mode of transmission remains unknown. In addition, recent studies suggest that this virus may latently infect the human sera and peripheral blood mononuclear cells.

Trichodysplasia spinulosa is a rare cutaneous condition that has been described almost exclusively in immunocompromised patients, usually organ transplant recipients, on regimens of immunosuppressive drugs. As of early 2016, a total of 32 cases had been reported in the medical literature. Despite its rarity, TS is believed to be underdiagnosed, and the growing population of patients on immunosuppressive drug regimens suggests its incidence may rise. TS has been described as an emerging infectious disease.

Murine polyomavirus is an unenveloped double-stranded DNA virus of the polyomavirus family. The first member of the family discovered, it was originally identified by accident in the 1950s. A component of mouse leukemia extract capable of causing tumors, particularly in the parotid gland, in newborn mice was reported by Ludwik Gross in 1953 and identified as a virus by Sarah Stewart and Bernice Eddy at the National Cancer Institute, after whom it was once called "SE polyoma". Stewart and Eddy would go on to study related polyomaviruses such as SV40 that infect primates, including humans. These discoveries were widely reported at the time and formed the early stages of understanding of oncoviruses.

Major capsid protein VP1 is a viral protein that is the main component of the polyomavirus capsid. VP1 monomers are generally around 350 amino acids long and are capable of self-assembly into an icosahedral structure consisting of 360 VP1 molecules organized into 72 pentamers. VP1 molecules possess a surface binding site that interacts with sialic acids attached to glycans, including some gangliosides, on the surfaces of cells to initiate the process of viral infection. The VP1 protein, along with capsid components VP2 and VP3, is expressed from the "late region" of the circular viral genome.
Hamster polyomavirus is an unenveloped double-stranded DNA virus of the polyomavirus family whose natural host is the hamster. It was originally described in 1967 by Arnold Graffi as a cause of epithelioma in Syrian hamsters.
WU polyomavirus is a virus of the family Polyomaviridae. It was discovered in 2007 in samples of human respiratory secretions, originally from a child patient in Australia who presented with clinical signs of pneumonia and in whom other common respiratory viruses were not detected. Follow-up studies identified the presence of WU virus in respiratory secretion samples from patients in Australia and the United States, suggesting that, like other human polyomaviruses, WU virus is widely distributed.
KI polyomavirus is a virus of the family Polyomaviridae. It was discovered in 2007 in stored samples of human respiratory secretions collected by the Karolinska Institute, after which the virus is named.

Agnoprotein is a protein expressed by some members of the polyomavirus family from a gene called the agnogene. Polyomaviruses in which it occurs include two human polyomaviruses associated with disease, BK virus and JC virus, as well as the simian polyomavirus SV40.
Human polyomavirus 7 (HPyV7) is a virus of the polyomavirus family that infects human hosts. It was discovered in 2010 and is a common component of the skin flora in healthy adults. There is limited evidence from case reports linking the virus to a skin rash occurring in immunocompromised organ transplant recipients.
Human polyomavirus 6 (HPyV6) is a virus of the polyomavirus family that infects human hosts. It was discovered in 2010 and is a component of the skin flora in healthy adults.
Human polyomavirus 9 (HPyV9) is a virus of the polyomavirus family that infects human hosts. It was discovered in 2011 and is a component of the skin flora in healthy adults.
Sorex araneus polyomavirus 1, formerly known as Human polyomavirus 12 (HPyV12), is a virus of the polyomavirus family that was first identified in human hosts and also infects shrews. It was discovered and reported in 2013 after isolation from the organs of the gastrointestinal tract, particularly the liver. The virus was renamed to Sorex araneus polyomavirus 1 in 2018, after discovery of the same virus in shrews. Infecting multiple hosts is rare among mammalian polyomaviruses.
MW polyomavirus is a virus of the polyomavirus family that infects human hosts. It was discovered in 2012 and reported independently by several research groups. It has been identified mostly in stool samples from children and has been detected in a variety of geographic locations.
STL polyomavirus is a virus of the polyomavirus family that infects human hosts. It was first reported in 2013 and is most closely related to MW polyomavirus. It has been identified mostly in stool samples from children and has been detected in a variety of geographic locations.
New Jersey polyomavirus is a virus of the polyomavirus family that infects human hosts. It was first identified in 2014 in a pancreatic transplant patient in New Jersey. It is the 13th and most recent human polyomavirus to be described.

The large tumor antigen is a protein encoded in the genomes of polyomaviruses, which are small double-stranded DNA viruses. LTag is expressed early in the infectious cycle and is essential for viral proliferation. Containing four well-conserved protein domains as well as several intrinsically disordered regions, LTag is a fairly large multifunctional protein; in most polyomaviruses, it ranges from around 600-800 amino acids in length. LTag has two primary functions, both related to replication of the viral genome: it unwinds the virus's DNA to prepare it for replication, and it interacts with proteins in the host cell to dysregulate the cell cycle so that the host's DNA replication machinery can be used to replicate the virus's genome. Some polyomavirus LTag proteins - most notably the well-studied SV40 large tumor antigen from the SV40 virus - are oncoproteins that can induce neoplastic transformation in the host cell.

The small tumor antigen is a protein encoded in the genomes of polyomaviruses, which are small double-stranded DNA viruses. STag is expressed early in the infectious cycle and is usually not essential for viral proliferation, though in most polyomaviruses it does improve replication efficiency. The STag protein is expressed from a gene that overlaps the large tumor antigen (LTag) such that the two proteins share an N-terminal DnaJ-like domain but have distinct C-terminal regions. STag is known to interact with host cell proteins, most notably protein phosphatase 2A (PP2A), and may activate the expression of cellular proteins associated with the cell cycle transition to S phase. In some polyomaviruses - such as the well-studied SV40, which natively infects monkeys - STag is unable to induce neoplastic transformation in the host cell on its own, but its presence may increase the transforming efficiency of LTag. In other polyomaviruses, such as Merkel cell polyomavirus, which causes Merkel cell carcinoma in humans, STag appears to be important for replication and to be an oncoprotein in its own right.
The middle tumor antigen is a protein encoded in the genomes of some polyomaviruses, which are small double-stranded DNA viruses. MTag is expressed early in the infectious cycle along with two other related proteins, the small tumor antigen and large tumor antigen. MTag occurs only in a few known polyomaviruses, while STag and LTag are universal - it was first identified in mouse polyomavirus (MPyV), the first polyomavirus discovered, and also occurs in hamster polyomavirus. In MPyV, MTag is an efficient oncoprotein that can be sufficient to induce neoplastic transformation in some cells.







