Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS and COVID-19, which is causing the ongoing pandemic. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis.

Severe acute respiratory syndrome–related coronavirus is a species of virus consisting of many known strains. Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1, which caused the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is causing the ongoing pandemic of COVID-19. There are hundreds of other strains of SARSr-CoV, which are only known to infect non-human mammal species: bats are a major reservoir of many strains of SARSr-CoV; several strains have been identified in Himalayan palm civets, which were likely ancestors of SARS-CoV-1.
Protease inhibitors (PIs) are medications that act by interfering with enzymes that cleave proteins. Some of the most well known are antiviral drugs widely used to treat HIV/AIDS, hepatitis C and COVID-19. These protease inhibitors prevent viral replication by selectively binding to viral proteases and blocking proteolytic cleavage of protein precursors that are necessary for the production of infectious viral particles.

Ritonavir, sold under the brand name Norvir, is an antiretroviral medication used along with other medications to treat HIV/AIDS. This combination treatment is known as highly active antiretroviral therapy (HAART). Ritonavir is a protease inhibitor and is used with other protease inhibitors. It may also be used in combination with other medications to treat hepatitis C and COVID-19. It is taken by mouth. Tablets of ritonavir are not bioequivalent to capsules, as the tablets may result in higher peak plasma concentrations.

Nidovirales is an order of enveloped, positive-strand RNA viruses which infect vertebrates and invertebrates. Host organisms include mammals, birds, reptiles, amphibians, fish, arthropods, molluscs, and helminths. The order includes the families Coronaviridae, Arteriviridae, Roniviridae, and Mesoniviridae.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo.
NS2-3 protease is an enzyme responsible for proteolytic cleavage between NS2 and NS3, which are non-structural proteins that form part of the HCV virus particle. NS3 protease of hepatitis C virus, on the other hand, is responsible for the cleavage of non-structural protein downstream. Both of these proteases are directly involved in HCV genome replication, that is, during the viral life-cycle that leads to virus multiplication in the host that has been infected by the virus.

Picornain 3C is a protease found in picornaviruses, which cleaves peptide bonds of non-terminal sequences. Picornain 3C’s endopeptidase activity is primarily responsible for the catalytic process of selectively cleaving Gln-Gly bonds in the polyprotein of poliovirus and with substitution of Glu for Gln, and Ser or Thr for Gly in other picornaviruses. Picornain 3C are cysteine proteases related by amino acid sequence to trypsin-like serine proteases. Picornain 3C is encoded by enteroviruses, rhinoviruses, aphtoviruses and cardioviruses. These genera of picoviruses cause a wide range of infections in humans and mammals.

Rupintrivir is a peptidomimetic antiviral drug which acts as a 3C and 3CL protease inhibitor. It was developed for the treatment of rhinoviruses, and has subsequently been investigated for the treatment of other viral diseases including those caused by picornaviruses, norovirus, and coronaviruses, such as SARS and COVID-19.

3CLpro-1 is an antiviral drug related to rupintrivir which acts as a 3CL protease inhibitor and was originally developed for the treatment of human enterovirus 71. It is one of the most potent of a large series of compounds developed as inhibitors of the viral enzyme 3CL protease, with an in vitroIC50 of 200 nM. It also shows activity against coronavirus diseases such as SARS and MERS, and is under investigation as a potential treatment agent for the viral disease COVID-19.

GC376 is a broad-spectrum antiviral medication under development by the biopharmaceutical company Anivive Lifesciences for therapeutic uses in humans and animals. Anivive licensed the exclusive worldwide patent rights to GC376 from Kansas State University. As of 2020, GC376 is being investigated as treatment for COVID-19. GC376 shows activity against many human and animal viruses including coronavirus and norovirus; the most extensive research has been multiple in vivo studies in cats treating a coronavirus which causes deadly feline infectious peritonitis. Other research supports use in porcine epidemic diarrhea virus.

Nirmatrelvir is an antiviral medication developed by Pfizer which acts as an orally active 3C-like protease inhibitor. It is part of a nirmatrelvir/ritonavir combination used to treat COVID-19 and sold under the brand name Paxlovid.

GRL-0617 is a drug which is one of the first compounds discovered that acts as a selective small-molecule inhibitor of the protease enzyme papain-like protease (PLpro) found in some pathogenic viruses, including the coronavirus SARS-CoV-2. It has been shown to inhibit viral replication in silico and in vitro.

Lufotrelvir (PF-07304814) is an antiviral drug developed by Pfizer which acts as a 3CL protease inhibitor. It is a prodrug with the phosphate group being cleaved in vivo to yield the active agent PF-00835231. Lufotrelvir is in human clinical trials for the treatment of COVID-19, and shows good activity against COVID-19 including several variant strains, but unlike the related drug nirmatrelvir it is not orally active and must be administered by intravenous infusion, and so has been the less favoured candidate for clinical development overall.
COVID Moonshot is a collaborative open-science project started in March 2020 with the goal of developing an un-patented oral antiviral drug to treat SARS-CoV-2, the virus causing COVID-19. COVID Moonshot researchers are targeting the proteins needed to form functioning new viral proteins. They are particularly interested in proteases such as 3C-like protease (Mpro), a coronavirus nonstructural protein that mediates the breaking and replication of proteins.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.

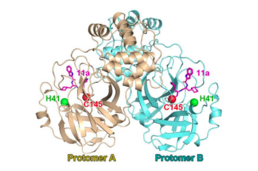
The nidoviral papain-like protease is a papain-like protease protein domain encoded in the genomes of nidoviruses. It is expressed as part of a large polyprotein from the ORF1a gene and has cysteine protease enzymatic activity responsible for proteolytic cleavage of some of the N-terminal viral nonstructural proteins within the polyprotein. A second protease also encoded by ORF1a, called the 3C-like protease or main protease, is responsible for the majority of further cleavages. Coronaviruses have one or two papain-like protease domains; in SARS-CoV and SARS-CoV-2, one PLPro domain is located in coronavirus nonstructural protein 3 (nsp3). Arteriviruses have two to three PLP domains. In addition to their protease activity, PLP domains function as deubiquitinating enzymes (DUBs) that can cleave the isopeptide bond found in ubiquitin chains. They are also "deISGylating" enzymes that remove the ubiquitin-like domain interferon-stimulated gene 15 (ISG15) from cellular proteins. These activities are likely responsible for antagonizing the activity of the host innate immune system. Because they are essential for viral replication, papain-like protease domains are considered drug targets for the development of antiviral drugs against human pathogens such as MERS-CoV, SARS-CoV, and SARS-CoV-2.

Papain-like proteases are a large protein family of cysteine protease enzymes that share structural and enzymatic properties with the group's namesake member, papain. They are found in all domains of life. In animals, the group is often known as cysteine cathepsins or, in older literature, lysosomal peptidases. In the MEROPS protease enzyme classification system, papain-like proteases form Clan CA. Papain-like proteases share a common catalytic dyad active site featuring a cysteine amino acid residue that acts as a nucleophile.

Ensitrelvir, sold under the brand name Xocova is an antiviral medication used as a treatment for COVID-19. It was developed by Shionogi in partnership with Hokkaido University and acts as an orally active 3C-like protease inhibitor. It is taken by mouth.

Olgotrelvir (STI-1558) is an experimental antiviral medication being studied to evaluate its potential as a treatment for COVID-19. It is believed to work by inhibiting the SARS-CoV-2 main protease (Mpro), a key enzyme the SARS-CoV-2 needs to replicate.