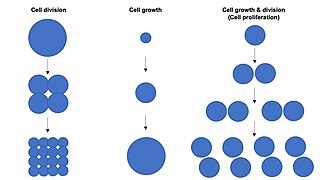
Cell growth refers to an increase in the total mass of a cell, including both cytoplasmic, nuclear and organelle volume. Cell growth occurs when the overall rate of cellular biosynthesis is greater than the overall rate of cellular degradation.

Autophagy is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly.

The mammalian target of rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the MTOR gene. mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases.
RHEB also known as Ras homolog enriched in brain (RHEB) is a GTP-binding protein that is ubiquitously expressed in humans and other mammals. The protein is largely involved in the mTOR pathway and the regulation of the cell cycle.

Ribosomal protein S6 kinase beta-1 (S6K1), also known as p70S6 kinase, is an enzyme that in humans is encoded by the RPS6KB1 gene. It is a serine/threonine kinase that acts downstream of PIP3 and phosphoinositide-dependent kinase-1 in the PI3 kinase pathway. As the name suggests, its target substrate is the S6 ribosomal protein. Phosphorylation of S6 induces protein synthesis at the ribosome.

Cell division cycle 7-related protein kinase is an enzyme that in humans is encoded by the CDC7 gene. The Cdc7 kinase is involved in regulation of the cell cycle at the point of chromosomal DNA replication. The gene CDC7 appears to be conserved throughout eukaryotic evolution; this means that most eukaryotic cells have the Cdc7 kinase protein.

Autophagy related 5 (ATG5) is a protein that, in humans, is encoded by the ATG5 gene located on Chromosome 6. It is an E3 ubi autophagic cell death. ATG5 is a key protein involved in the extension of the phagophoric membrane in autophagic vesicles. It is activated by ATG7 and forms a complex with ATG12 and ATG16L1. This complex is necessary for LC3-I conjugation to PE (phosphatidylethanolamine) to form LC3-II. ATG5 can also act as a pro-apoptotic molecule targeted to the mitochondria. Under low levels of DNA damage, ATG5 can translocate to the nucleus and interact with survivin.

Regulatory-associated protein of mTOR also known as raptor or KIAA1303 is an adapter protein that is encoded in humans by the RPTOR gene. Two mRNAs from the gene have been identified that encode proteins of 1335 and 1177 amino acids long.

Autophagy related 12 is a protein that in humans is encoded by the ATG12 gene.

ULK1 is an enzyme that in humans is encoded by the ULK1 gene.

Autophagy related 7 is a protein in humans encoded by ATG7 gene. Related to GSA7; APG7L; APG7-LIKE.
The Akt signaling pathway or PI3K-Akt signaling pathway is a signal transduction pathway that promotes survival and growth in response to extracellular signals. Key proteins involved are PI3K and Akt.

Autophagy-related protein 8 (Atg8) is a ubiquitin-like protein required for the formation of autophagosomal membranes. The transient conjugation of Atg8 to the autophagosomal membrane through a ubiquitin-like conjugation system is essential for autophagy in eukaryotes. Even though there are homologues in animals, this article mainly focuses on its role in lower eukaryotes such as Saccharomyces cerevisiae.

Autophagy-related protein 13 also known as ATG13 is a protein that in humans is encoded by the KIAA0652 gene.
An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents. After formation, autophagosomes deliver cytoplasmic components to the lysosomes. The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome's hydrolases degrade the autophagosome-delivered contents and its inner membrane.
In molecular biology, autophagy related 3 (Atg3) is the E2 enzyme for the LC3 lipidation process. It is essential for autophagy. The super protein complex, the Atg16L complex, consists of multiple Atg12-Atg5 conjugates. Atg16L has an E3-like role in the LC3 lipidation reaction. The activated intermediate, LC3-Atg3 (E2), is recruited to the site where the lipidation takes place.

Target of rapamycin complex subunit LST8, also known as mammalian lethal with SEC13 protein 8 (mLST8) or TORC subunit LST8 or G protein beta subunit-like, is a protein that in humans is encoded by the MLST8 gene. It is a subunit of both mTORC1 and mTORC2, complexes that regulate cell growth and survival in response to nutrient, energy, redox, and hormonal signals. It is upregulated in several human colon and prostate cancer cell lines and tissues. Knockdown of mLST8 prevented mTORC formation and inhibited tumor growth and invasiveness.

Yoshinori Ohsumi is a Japanese cell biologist specializing in autophagy, the process that cells use to destroy and recycle cellular components. Ohsumi is a professor at Tokyo Institute of Technology's Institute of Innovative Research. He received the Kyoto Prize for Basic Sciences in 2012, the 2016 Nobel Prize in Physiology or Medicine, and the 2017 Breakthrough Prize in Life Sciences for his discoveries of mechanisms for autophagy.

mTORC1, also known as mammalian target of rapamycin complex 1 or mechanistic target of rapamycin complex 1, is a protein complex that functions as a nutrient/energy/redox sensor and controls protein synthesis.

The Ragulator-Rag complex is a regulator of lysosomal signalling and trafficking in eukaryotic cells, which plays an important role in regulating cell metabolism and growth in response to nutrient availability in the cell. The Ragulator-Rag Complex is composed of five LAMTOR subunits, which work to regulate MAPK and mTOR complex 1. The LAMTOR subunits form a complex with Rag GTPase and v-ATPase, which sits on the cell’s lysosomes and detects the availability of amino acids. If the Ragulator complex receives signals for low amino acid count, it will start the process of catabolizing the cell. If there is an abundance of amino acids available to the cell, the Ragulator complex will signal that the cell can continue to grow. Ragulator proteins come in two different forms: Rag A/Rag B and Rag C/Rag D. These interact to form heterodimers with one another.



















