Cladistics is an approach to biological classification in which organisms are categorized in groups ("clades") based on hypotheses of most recent common ancestry. The evidence for hypothesized relationships is typically shared derived characteristics (synapomorphies) that are not present in more distant groups and ancestors. However, from an empirical perspective, common ancestors are inferences based on a cladistic hypothesis of relationships of taxa whose character states can be observed. Theoretically, a last common ancestor and all its descendants constitute a (minimal) clade. Importantly, all descendants stay in their overarching ancestral clade. For example, if the terms worms or fishes were used within a strict cladistic framework, these terms would include humans. Many of these terms are normally used paraphyletically, outside of cladistics, e.g. as a 'grade', which are fruitless to precisely delineate, especially when including extinct species. Radiation results in the generation of new subclades by bifurcation, but in practice sexual hybridization may blur very closely related groupings.
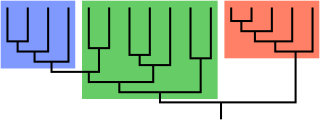
In biological phylogenetics, a clade, also known as a monophyletic group or natural group, is a grouping of organisms that are monophyletic – that is, composed of a common ancestor and all its lineal descendants – on a phylogenetic tree. In the taxonomical literature, sometimes the Latin form cladus is used rather than the English form. Clades are the fundamental unit of cladistics, a modern approach to taxonomy adopted by most biological fields.
In biology, phylogenetics is the study of the evolutionary history and relationships among or within groups of organisms. These relationships are determined by phylogenetic inference methods that focus on observed heritable traits, such as DNA sequences, protein amino acid sequences, or morphology. The result of such an analysis is a phylogenetic tree—a diagram containing a hypothesis of relationships that reflects the evolutionary history of a group of organisms.

Paraphyly is a taxonomic term describing a grouping that consists of the grouping's last common ancestor and most of its descendants, but excludes one or more subgroups. The grouping is said to be paraphyletic with respect to the excluded subgroups. In contrast, a monophyletic grouping includes a common ancestor and all of its descendants.

A cladogram is a diagram used in cladistics to show relations among organisms. A cladogram is not, however, an evolutionary tree because it does not show how ancestors are related to descendants, nor does it show how much they have changed, so many differing evolutionary trees can be consistent with the same cladogram. A cladogram uses lines that branch off in different directions ending at a clade, a group of organisms with a last common ancestor. There are many shapes of cladograms but they all have lines that branch off from other lines. The lines can be traced back to where they branch off. These branching off points represent a hypothetical ancestor which can be inferred to exhibit the traits shared among the terminal taxa above it. This hypothetical ancestor might then provide clues about the order of evolution of various features, adaptation, and other evolutionary narratives about ancestors. Although traditionally such cladograms were generated largely on the basis of morphological characters, DNA and RNA sequencing data and computational phylogenetics are now very commonly used in the generation of cladograms, either on their own or in combination with morphology.

Evolutionary developmental biology is a field of biological research that compares the developmental processes of different organisms to infer how developmental processes evolved.

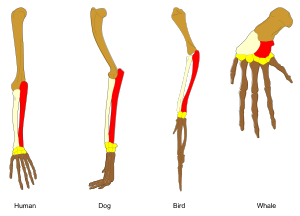
Convergent evolution is the independent evolution of similar features in species of different periods or epochs in time. Convergent evolution creates analogous structures that have similar form or function but were not present in the last common ancestor of those groups. The cladistic term for the same phenomenon is homoplasy. The recurrent evolution of flight is a classic example, as flying insects, birds, pterosaurs, and bats have independently evolved the useful capacity of flight. Functionally similar features that have arisen through convergent evolution are analogous, whereas homologous structures or traits have a common origin but can have dissimilar functions. Bird, bat, and pterosaur wings are analogous structures, but their forelimbs are homologous, sharing an ancestral state despite serving different functions.
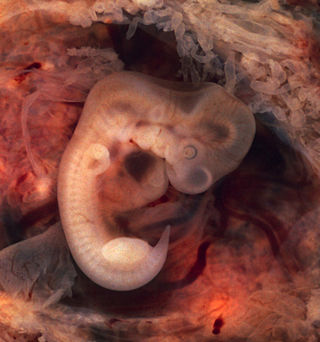
In biology, an atavism is a modification of a biological structure whereby an ancestral genetic trait reappears after having been lost through evolutionary change in previous generations. Atavisms can occur in several ways, one of which is when genes for previously existing phenotypic features are preserved in DNA, and these become expressed through a mutation that either knocks out the dominant genes for the new traits or makes the old traits dominate the new one. A number of traits can vary as a result of shortening of the fetal development of a trait (neoteny) or by prolongation of the same. In such a case, a shift in the time a trait is allowed to develop before it is fixed can bring forth an ancestral phenotype. Atavisms are often seen as evidence of evolution.

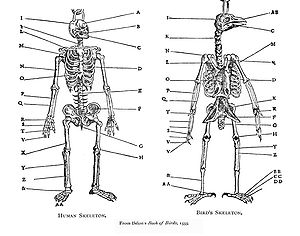
Comparative anatomy is the study of similarities and differences in the anatomy of different species. It is closely related to evolutionary biology and phylogeny.

Vestigiality is the retention, during the process of evolution, of genetically determined structures or attributes that have lost some or all of the ancestral function in a given species. Assessment of the vestigiality must generally rely on comparison with homologous features in related species. The emergence of vestigiality occurs by normal evolutionary processes, typically by loss of function of a feature that is no longer subject to positive selection pressures when it loses its value in a changing environment. The feature may be selected against more urgently when its function becomes definitively harmful, but if the lack of the feature provides no advantage, and its presence provides no disadvantage, the feature may not be phased out by natural selection and persist across species.

A polyphyletic group is an assemblage that includes organisms with mixed evolutionary origin but does not include their most recent common ancestor. The term is often applied to groups that share similar features known as homoplasies, which are explained as a result of convergent evolution. The arrangement of the members of a polyphyletic group is called a polyphyly. It is contrasted with monophyly and paraphyly.
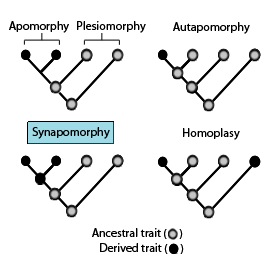
In phylogenetics, an apomorphy is a novel character or character state that has evolved from its ancestral form. A synapomorphy is an apomorphy shared by two or more taxa and is therefore hypothesized to have evolved in their most recent common ancestor. In cladistics, synapomorphy implies homology.

Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal gene transfer event (xenologs).
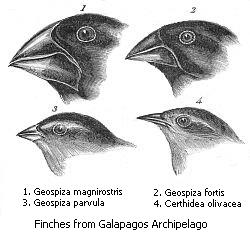
Divergent evolution or divergent selection is the accumulation of differences between closely related populations within a species, sometimes leading to speciation. Divergent evolution is typically exhibited when two populations become separated by a geographic barrier and experience different selective pressures that drive adaptations to their new environment. After many generations and continual evolution, the populations become less able to interbreed with one another. The American naturalist J. T. Gulick (1832–1923) was the first to use the term "divergent evolution", with its use becoming widespread in modern evolutionary literature. Classic examples of divergence in nature are the adaptive radiation of the finches of the Galapagos or the coloration differences in populations of a species that live in different habitats such as with pocket mice and fence lizards.
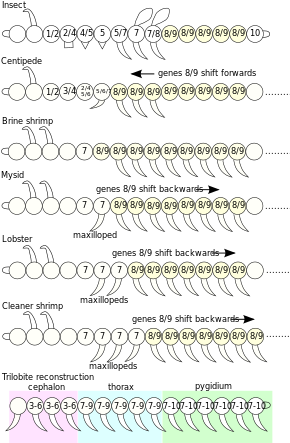
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment, and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves.

In phylogenetics, a plesiomorphy and symplesiomorphy are synonyms for an ancestral character shared by all members of a clade, which does not distinguish the clade from other clades.
The arthropod leg is a form of jointed appendage of arthropods, usually used for walking. Many of the terms used for arthropod leg segments are of Latin origin, and may be confused with terms for bones: coxa, trochanter, femur, tibia, tarsus, ischium, metatarsus, carpus, dactylus, patella.
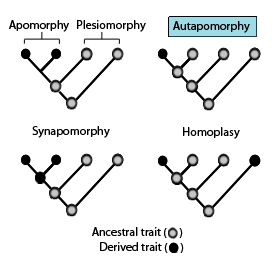
In phylogenetics, an autapomorphy is a distinctive feature, known as a derived trait, that is unique to a given taxon. That is, it is found only in one taxon, but not found in any others or outgroup taxa, not even those most closely related to the focal taxon. It can therefore be considered an apomorphy in relation to a single taxon. The word autapomorphy, introduced in 1950 by German entomologist Willi Hennig, is derived from the Greek words αὐτός, autos "self"; ἀπό, apo "away from"; and μορφή, morphḗ = "shape".

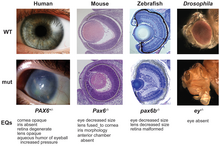
In evolutionary developmental biology, the concept of deep homology is used to describe cases where growth and differentiation processes are governed by genetic mechanisms that are homologous and deeply conserved across a wide range of species.
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred. Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent. Superfamilies typically contain several protein families which show sequence similarity within each family. The term protein clan is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems.