The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.

In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements.

In molecular biology, a stop codon is a codon that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.
Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, the evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.

In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression.
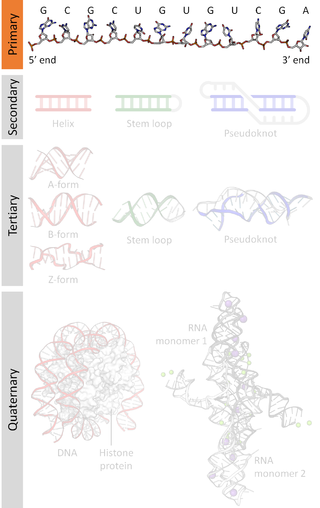
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a nonsense codon, or a premature stop codon in the transcribed mRNA, and leads to a truncated, incomplete, and possibly nonfunctional protein product. Nonsense mutations are not always harmful; the functional effect of a nonsense mutation depends on many aspects, such as the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations.

Silent mutations are mutations in DNA that do not have an observable effect on the organism's phenotype. They are a specific type of neutral mutation. The phrase silent mutation is often used interchangeably with the phrase synonymous mutation; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually significantly affect protein function.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.
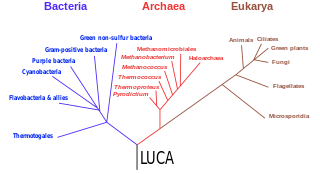
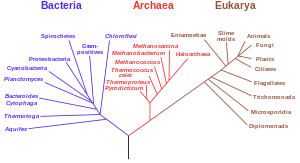
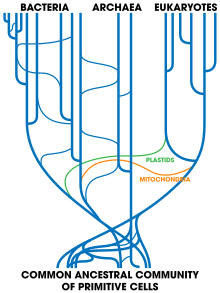
The last universal common ancestor (LUCA) is the hypothesized common ancestral cell from which the three domains of life, the Bacteria, the Archaea, and the Eukarya originated. It is suggested to have been a "cellular organism that had a lipid bilayer and used DNA, RNA, and protein". The LUCA has also been defined as "a hypothetical organism ancestral to all three domains". The LUCA is the point or stage at which the three domains of life diverged from preexisting forms of life. The nature of this point or stage of divergence remains a topic of research.
Evidence of common descent of living organisms has been discovered by scientists researching in a variety of disciplines over many decades, demonstrating that all life on Earth comes from a single ancestor. This forms an important part of the evidence on which evolutionary theory rests, demonstrates that evolution does occur, and illustrates the processes that created Earth's biodiversity. It supports the modern evolutionary synthesis—the current scientific theory that explains how and why life changes over time. Evolutionary biologists document evidence of common descent, all the way back to the last universal common ancestor, by developing testable predictions, testing hypotheses, and constructing theories that illustrate and describe its causes.

In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA, that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes.
Steven Albert Benner is an American chemist. He has been a professor at Harvard University, ETH Zurich, and most recently at the University of Florida, where he was the V.T. & Louise Jackson Distinguished Professor of Chemistry. In 2005, he founded The Westheimer Institute of Science and Technology (TWIST) and the Foundation For Applied Molecular Evolution. Benner has also founded the companies EraGen Biosciences and Firebird BioMolecular Sciences LLC.
Neutral mutations are changes in DNA sequence that are neither beneficial nor detrimental to the ability of an organism to survive and reproduce. In population genetics, mutations in which natural selection does not affect the spread of the mutation in a species are termed neutral mutations. Neutral mutations that are inheritable and not linked to any genes under selection will be lost or will replace all other alleles of the gene. That loss or fixation of the gene proceeds based on random sampling known as genetic drift. A neutral mutation that is in linkage disequilibrium with other alleles that are under selection may proceed to loss or fixation via genetic hitchhiking and/or background selection.

An expanded genetic code is an artificially modified genetic code in which one or more specific codons have been re-allocated to encode an amino acid that is not among the 22 common naturally-encoded proteinogenic amino acids.

An organism is any biological living system that functions as an individual life form. All organisms are composed of cells. The idea of organism is based on the concept of minimal functional unit of life. Three traits have been proposed to play the main role in qualification as an organism:
Evolution of cells refers to the evolutionary origin and subsequent evolutionary development of cells. Cells first emerged at least 3.8 billion years ago approximately 750 million years after Earth was formed.

A codon table can be used to translate a genetic code into a sequence of amino acids. The standard genetic code is traditionally represented as an RNA codon table, because when proteins are made in a cell by ribosomes, it is messenger RNA (mRNA) that directs protein synthesis. The mRNA sequence is determined by the sequence of genomic DNA. In this context, the standard genetic code is referred to as translation table 1. It can also be represented in a DNA codon table. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5′-to-3′ direction. Different tables with alternate codons are used depending on the source of the genetic code, such as from a cell nucleus, mitochondrion, plastid, or hydrogenosome.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution, and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.
The split gene theory is a theory of the origin of introns, long non-coding sequences in eukaryotic genes between the exons. The theory holds that the randomness of primordial DNA sequences would only permit small (< 600bp) open reading frames (ORFs), and that important intron structures and regulatory sequences are derived from stop codons. In this introns-first framework, the spliceosomal machinery and the nucleus evolved due to the necessity to join these ORFs into larger proteins, and that intronless bacterial genes are less ancestral than the split eukaryotic genes. The theory originated with Periannan Senapathy.