
The Mexican tetra, also known as the blind cave fish, blind cave characin, and blind cave tetra, is a freshwater fish of the family Characidae of the order Characiformes. The type species of its genus, it is native to the Nearctic realm, originating in the lower Rio Grande and the Neueces and Pecos Rivers in Texas, as well as the central and eastern parts of Mexico.

Evolution is the change in the heritable characteristics of biological populations over successive generations. Evolution occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations. The process of evolution has given rise to biodiversity at every level of biological organisation.
Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.

Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Charles Darwin popularised the term "natural selection", contrasting it with artificial selection, which is intentional, whereas natural selection is not.

In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems by relying on biologically inspired operators such as mutation, crossover and selection. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, causal inference, etc.
Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, the evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.

In computational intelligence (CI), an evolutionary algorithm (EA) is a subset of evolutionary computation, a generic population-based metaheuristic optimization algorithm. An EA uses mechanisms inspired by biological evolution, such as reproduction, mutation, recombination, and selection. Candidate solutions to the optimization problem play the role of individuals in a population, and the fitness function determines the quality of the solutions. Evolution of the population then takes place after the repeated application of the above operators.
In computer science, evolutionary computation is a family of algorithms for global optimization inspired by biological evolution, and the subfield of artificial intelligence and soft computing studying these algorithms. In technical terms, they are a family of population-based trial and error problem solvers with a metaheuristic or stochastic optimization character.

Evolutionary biology is the subfield of biology that studies the evolutionary processes that produced the diversity of life on Earth. It is also defined as the study of the history of life forms on Earth. Evolution holds that all species are related and gradually change over generations. In a population, the genetic variations affect the phenotypes of an organism. These changes in the phenotypes will be an advantage to some organisms, which will then be passed onto their offspring. Some examples of evolution in species over many generations are the peppered moth and flightless birds. In the 1930s, the discipline of evolutionary biology emerged through what Julian Huxley called the modern synthesis of understanding, from previously unrelated fields of biological research, such as genetics and ecology, systematics, and paleontology.
Neuroevolution, or neuro-evolution, is a form of artificial intelligence that uses evolutionary algorithms to generate artificial neural networks (ANN), parameters, and rules. It is most commonly applied in artificial life, general game playing and evolutionary robotics. The main benefit is that neuroevolution can be applied more widely than supervised learning algorithms, which require a syllabus of correct input-output pairs. In contrast, neuroevolution requires only a measure of a network's performance at a task. For example, the outcome of a game can be easily measured without providing labeled examples of desired strategies. Neuroevolution is commonly used as part of the reinforcement learning paradigm, and it can be contrasted with conventional deep learning techniques that use gradient descent on a neural network with a fixed topology.
Experimental evolution is the use of laboratory experiments or controlled field manipulations to explore evolutionary dynamics. Evolution may be observed in the laboratory as individuals/populations adapt to new environmental conditions by natural selection.
Evolvability is defined as the capacity of a system for adaptive evolution. Evolvability is the ability of a population of organisms to not merely generate genetic diversity, but to generate adaptive genetic diversity, and thereby evolve through natural selection.
In computer science, an evolution strategy (ES) is an optimization technique based on ideas of evolution. It belongs to the general class of evolutionary computation or artificial evolution methodologies.
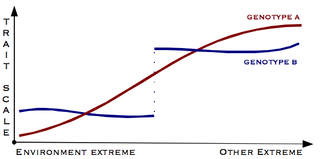
Canalisation is a measure of the ability of a population to produce the same phenotype regardless of variability of its environment or genotype. It is a form of evolutionary robustness. The term was coined in 1942 by C. H. Waddington to capture the fact that "developmental reactions, as they occur in organisms submitted to natural selection...are adjusted so as to bring about one definite end-result regardless of minor variations in conditions during the course of the reaction". He used this word rather than robustness to consider that biological systems are not robust in quite the same way as, for example, engineered systems.
Enquiry into the evolution of ageing, or aging, aims to explain why a detrimental process such as ageing would evolve, and why there is so much variability in the lifespans of organisms. The classical theories of evolution suggest that environmental factors, such as predation, accidents, disease, and/or starvation, ensure that most organisms living in natural settings will not live until old age, and so there will be very little pressure to conserve genetic changes that increase longevity. Natural selection will instead strongly favor genes which ensure early maturation and rapid reproduction, and the selection for genetic traits which promote molecular and cellular self-maintenance will decline with age for most organisms.

Clonal interference is a phenomenon in evolutionary biology, related to the population genetics of organisms with significant linkage disequilibrium, especially asexually reproducing organisms. The idea of clonal interference was introduced by American geneticist Hermann Joseph Muller in 1932. It explains why beneficial mutations can take a long time to get fixated or even disappear in asexually reproducing populations. As the name suggests, clonal interference occurs in an asexual lineage ("clone") with a beneficial mutation. This mutation would be likely to get fixed if it occurred alone, but it may fail to be fixed, or even be lost, if another beneficial-mutation lineage arises in the same population; the multiple clones interfere with each other.
Natural computing, also called natural computation, is a terminology introduced to encompass three classes of methods: 1) those that take inspiration from nature for the development of novel problem-solving techniques; 2) those that are based on the use of computers to synthesize natural phenomena; and 3) those that employ natural materials to compute. The main fields of research that compose these three branches are artificial neural networks, evolutionary algorithms, swarm intelligence, artificial immune systems, fractal geometry, artificial life, DNA computing, and quantum computing, among others.

In evolutionary biology, robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.

The following outline is provided as an overview of and topical guide to evolution:

Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dependent on the genetic background in which it appears. Epistatic mutations therefore have different effects on their own than when they occur together. Originally, the term epistasis specifically meant that the effect of a gene variant is masked by that of different gene.












