A pilus is a hair-like appendage found on the surface of many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric.
Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
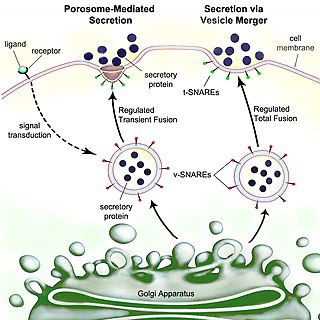
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.
Adhesins are cell-surface components or appendages of bacteria that facilitate adhesion or adherence to other cells or to surfaces, usually in the host they are infecting or living in. Adhesins are a type of virulence factor.
Pilin refers to a class of fibrous proteins that are found in pilus structures in bacteria. These structures can be used for the exchange of genetic material, or as a cell adhesion mechanism. Although not all bacteria have pili or fimbriae, bacterial pathogens often use their fimbriae to attach to host cells. In Gram-negative bacteria, where pili are more common, individual pilin molecules are linked by noncovalent protein-protein interactions, while Gram-positive bacteria often have polymerized LPXTG pilin.

Mitochondrial membrane transport proteins, also known as mitochondrial carrier proteins, are proteins which exist in the membranes of mitochondria. They serve to transport molecules and other factors, such as ions, into or out of the organelles. Mitochondria contain both an inner and outer membrane, separated by the inter-membrane space, or inner boundary membrane. The outer membrane is porous, whereas the inner membrane restricts the movement of all molecules. The two membranes also vary in membrane potential and pH. These factors play a role in the function of mitochondrial membrane transport proteins. There are 53 discovered human mitochondrial membrane transporters, with many others that are known to still need discovered.
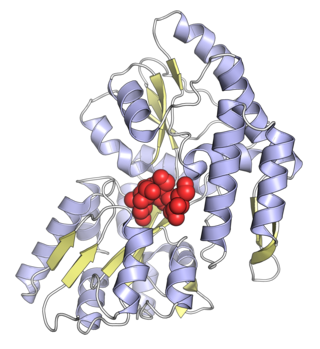
Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of Escherichia coli, which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many proteins and protein complexes. MBP has an approximate molecular mass of 42.5 kilodaltons.
Porphyromonas gingivalis belongs to the phylum Bacteroidota and is a nonmotile, Gram-negative, rod-shaped, anaerobic, pathogenic bacterium. It forms black colonies on blood agar.
In biology, phase variation is a method for dealing with rapidly varying environments without requiring random mutation. It involves the variation of protein expression, frequently in an on-off fashion, within different parts of a bacterial population. As such the phenotype can switch at frequencies that are much higher than classical mutation rates. Phase variation contributes to virulence by generating heterogeneity. Although it has been most commonly studied in the context of immune evasion, it is observed in many other areas as well and is employed by various types of bacteria, including Salmonella species.
Sortase refers to a group of prokaryotic enzymes that modify surface proteins by recognizing and cleaving a carboxyl-terminal sorting signal. For most substrates of sortase enzymes, the recognition signal consists of the motif LPXTG (Leu-Pro-any-Thr-Gly), then a highly hydrophobic transmembrane sequence, followed by a cluster of basic residues such as arginine. Cleavage occurs between the Thr and Gly, with transient attachment through the Thr residue to the active site Cys residue, followed by transpeptidation that attaches the protein covalently to cell wall components. Sortases occur in almost all Gram-positive bacteria and the occasional Gram-negative bacterium or Archaea, where cell wall LPXTG-mediated decoration has not been reported. Although sortase A, the "housekeeping" sortase, typically acts on many protein targets, other forms of sortase recognize variant forms of the cleavage motif, or catalyze the assembly of pilins into pili.
The PilZ protein family is named after the type IV pilus control protein first identified in Pseudomonas aeruginosa, expressed as part of the pil operon. It has a cytoplasmic location and is essential for type IV fimbrial, or pilus, biogenesis. PilZ is a c-di-GMP binding domain and PilZ domain-containing proteins represent the best studied class of c-di-GMP effectors. C-di-GMP, cyclic diguanosine monophosphate, the second messenger in cells, is widespread in and unique to the bacterial kingdom. Elevated intracellular levels of c-di-GMP generally cause bacteria to change from a motile single-cell state to a sessile, adhesive surface-attached multicellular state called biofilm.

In molecular biology, trimeric autotransporter adhesins (TAAs), are proteins found on the outer membrane of Gram-negative bacteria. Bacteria use TAAs in order to infect their host cells via a process called cell adhesion. TAAs also go by another name, oligomeric coiled-coil adhesins, which is shortened to OCAs. In essence, they are virulence factors, factors that make the bacteria harmful and infective to the host organism.
Chaperone-usher fimbriae (CU) are linear, unbranching, outer-membrane pili secreted by gram-negative bacteria through the chaperone-usher system rather than through type IV secretion or extracellular nucleation systems. These fimbriae are built up out of modular pilus subunits, which are transported into the periplasm in a Sec dependent manner. Chaperone-usher secreted fimbriae are important pathogenicity factors facilitating host colonisation, localisation and biofilm formation in clinically important species such as uropathogenic Escherichia coli and Pseudomonas aeruginosa.

Outer membrane vesicles (OMVs) are vesicles released from the outer membranes of Gram-negative bacteria. While Gram-positive bacteria release vesicles as well those vesicles fall under the broader category of bacterial membrane vesicles (MVs). OMVs were the first MVs to be discovered, and are distinguished from outer inner membrane vesicles (OIMVS), which are gram-negative baterial vesicles containing portions of both the outer and inner bacterial membrane. Outer membrane vesicles were first discovered and characterized using transmission-electron microscopy by Indian Scientist Prof. Smriti Narayan Chatterjee and J. Das in 1966-67. OMVs are ascribed the functionality to provide a manner to communicate among themselves, with other microorganisms in their environment and with the host. These vesicles are involved in trafficking bacterial cell signaling biochemicals, which may include DNA, RNA, proteins, endotoxins and allied virulence molecules. This communication happens in microbial cultures in oceans, inside animals, plants and even inside the human body.
Gabriel Waksman FMedSci, FRS, is Courtauld professor of biochemistry and molecular biology at University College London (UCL), and professor of structural and molecular biology at Birkbeck College, University of London. He is the director of the Institute of Structural and Molecular Biology (ISMB) at UCL and Birkbeck, head of the Department of Structural and Molecular Biology at UCL, and head of the Department of Biological Sciences at Birkbeck.
The type 2 secretion system is a type of protein secretion machinery found in various species of Gram-negative bacteria, including many human pathogens such as Pseudomonas aeruginosa and Vibrio cholerae. The type II secretion system is one of six protein secretory systems commonly found in Gram-negative bacteria, along with the type I, type III, and type IV secretion systems, as well as the chaperone/usher pathway, the autotransporter pathway/type V secretion system, and the type VI secretion system. Like these other systems, the type II secretion system enables the transport of cytoplasmic proteins across the lipid bilayers that make up the cell membranes of Gram-negative bacteria. Secretion of proteins and effector molecules out of the cell plays a critical role in signaling other cells and in the invasion and parasitism of host cells.
BamA is a β-barrel, outer membrane protein found in Gram-negative bacteria and it is the main and vital component of the β-barrel assembly machinery (BAM) complex in those bacteria. BAM Complex consists of five components; BamB, BamC, BamD, BamE and BamA. This complex is responsible in catalyzing folding and insertion of β-barrel proteins into the outer membrane of Gram-negative bacteria.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.
P fimbriae are chaperone-usher type fimbrial appendages found on the surface of many Escherichia coli bacteria. The P fimbriae is considered to be one of the most important virulence factor in uropathogenic E. coli and plays an important role in upper urinary tract infections. P fimbriae mediate adherence to host cells, a key event in the pathogenesis of urinary tract infections.
Type VII secretion systems are bacterial secretion systems first observed in the phyla Actinomycetota and Bacillota. Bacteria use such systems to transport, or secrete, proteins into the environment. The bacterial genus Mycobacterium uses type VII secretion systems (T7SS) to secrete proteins across their cell envelope. The first T7SS system discovered was the ESX-1 System.