
The proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. Proteomics is the study of the proteome.

Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.
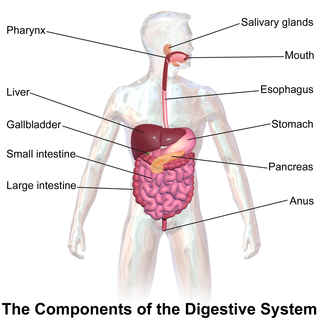
Digestion is the breakdown of large insoluble food molecules into small water-soluble food molecules so that they can be absorbed into the watery blood plasma. In certain organisms, these smaller substances are absorbed through the small intestine into the blood stream. Digestion is a form of catabolism that is often divided into two processes based on how food is broken down: mechanical and chemical digestion. The term mechanical digestion refers to the physical breakdown of large pieces of food into smaller pieces which can subsequently be accessed by digestive enzymes. Mechanical digestion takes place in the mouth through mastication and in the small intestine through segmentation contractions. In chemical digestion, enzymes break down food into the small molecules the body can use.

Food science is the basic science and applied science of food; its scope starts at overlap with agricultural science and nutritional science and leads through the scientific aspects of food safety and food processing, informing the development of food technology.

Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach to biological research.
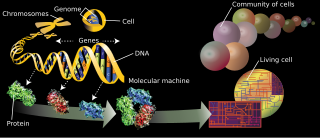
The branches of science known informally as omics are various disciplines in biology whose names end in the suffix -omics, such as genomics, proteomics, metabolomics, metagenomics, phenomics and transcriptomics. Omics aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms.

Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates, and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles. The metabolome represents the complete set of metabolites in a biological cell, tissue, organ, or organism, which are the end products of cellular processes. Messenger RNA (mRNA), gene expression data, and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell, and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism. There are indeed quantifiable correlations between the metabolome and the other cellular ensembles, which can be used to predict metabolite abundances in biological samples from, for example mRNA abundances. One of the ultimate challenges of systems biology is to integrate metabolomics with all other -omics information to provide a better understanding of cellular biology.
Nutritional genomics, also known as nutrigenomics, is a science studying the relationship between human genome, human nutrition and health. People in the field work toward developing an understanding of how the whole body responds to a food via systems biology, as well as single gene/single food compound relationships. Nutritional genomics or Nutrigenomics is the relation between food and inherited genes, it was first expressed in 2001.
This page provides an alphabetical list of articles and other pages about biotechnology.

In pharmacology, the term mechanism of action (MOA) refers to the specific biochemical interaction through which a drug substance produces its pharmacological effect. A mechanism of action usually includes mention of the specific molecular targets to which the drug binds, such as an enzyme or receptor. Receptor sites have specific affinities for drugs based on the chemical structure of the drug, as well as the specific action that occurs there.
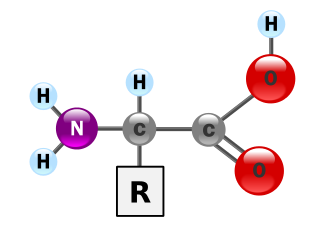
Proteins are essential nutrients for the human body. They are one of the building blocks of body tissue and can also serve as a fuel source. As a fuel, proteins provide as much energy density as carbohydrates: 4 kcal per gram; in contrast, lipids provide 9 kcal per gram. The most important aspect and defining characteristic of protein from a nutritional standpoint is its amino acid composition.

Proteogenomics is a field of biological research that utilizes a combination of proteomics, genomics, and transcriptomics to aid in the discovery and identification of peptides. Proteogenomics is used to identify new peptides by comparing MS/MS spectra against a protein database that has been derived from genomic and transcriptomic information. Proteogenomics often refers to studies that use proteomic information, often derived from mass spectrometry, to improve gene annotations. The utilization of both proteomics and genomics data alongside advances in the availability and power of spectrographic and chromatographic technology led to the emergence of proteogenomics as its own field in 2004.
The Human Protein Atlas (HPA) is a Swedish-based program started in 2003 with the aim to map all the human proteins in cells, tissues and organs using integration of various omics technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics and systems biology. All the data in the knowledge resource is open access to allow scientists both in academia and industry to freely access the data for exploration of the human proteome. In December 2022, version 22 was launched where two new sections, a Human Disease Blood Atlas and a Structure resource section, were introduced, both relying heavily on AI-based prediction modelling and machine learning.

In the field of cellular biology, single-cell analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell in a bulk population of cells at a particular condition using optical or electronic microscope. To date, due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome and transcriptome of single cells, as well as to quantify their proteome and metabolome. Mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. Recent advances have enabled quantifying thousands of protein across hundreds of single cells, and thus make possible new types of analysis. In situ sequencing and fluorescence in situ hybridization (FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.
Pan-cancer analysis aims to examine the similarities and differences among the genomic and cellular alterations found across diverse tumor types. International efforts have performed pan-cancer analysis on exomes and the whole genomes of cancers, the latter including their non-coding regions. In 2018, The Cancer Genome Atlas (TCGA) Research Network used exome, transcriptome, and DNA methylome data to develop an integrated picture of commonalities, differences, and emergent themes across tumor types.
Volatolomics is a branch of chemistry that studies volatile organic compounds (VOCs) emitted by a biological system, under specific experimental conditions.

Multiomics, multi-omics, integrative omics, "panomics" or "pan-omics" is a biological analysis approach in which the data sets are multiple "omes", such as the genome, proteome, transcriptome, epigenome, metabolome, and microbiome ; in other words, the use of multiple omics technologies to study life in a concerted way. By combining these "omes", scientists can analyze complex biological big data to find novel associations between biological entities, pinpoint relevant biomarkers and build elaborate markers of disease and physiology. In doing so, multiomics integrates diverse omics data to find a coherently matching geno-pheno-envirotype relationship or association. The OmicTools service lists more than 99 softwares related to multiomic data analysis, as well as more than 99 databases on the topic.
Michael P. Snyder is an American genomicist who is the Stanford B. Ascherman Professor and as of 2009, chair of genetics and director of genomics and personalized medicine at Stanford University, and the former director of the Yale Center for Genomics and Proteomics. He was elected to the American Academy of Arts and Sciences in 2015. During his tenure as chair of the department at Stanford, U.S. News & World Report has ranked Stanford University first or tied for first in genetics, genomics and bioinformatics under his leadership.
Deterministic Barcoding in Tissue for Spatial Omics Sequencing (DBiT-seq) was developed at Yale University by Rong Fan and colleagues in 2020 to create a multi-omics approach for studying spatial gene expression heterogenicity within a tissue sample. This method can used for the co-mapping mRNA and protein levels at a near single-cell resolution in fresh or frozen formaldehyde-fixed tissue samples. DBiT-seq utilizes next generation sequencing (NGS) and microfluidics. This method allows for simultaneous spatial transcriptomic and proteomic analysis of a tissue sample. DBiT-seq improves upon previous spatial transcriptomics applications such as High-Definition Spatial Transcriptomics (HDST) and Slide-seq by increasing the number of detectable genes per pixel, increased cellular resolution, and ease of implementation.














