A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
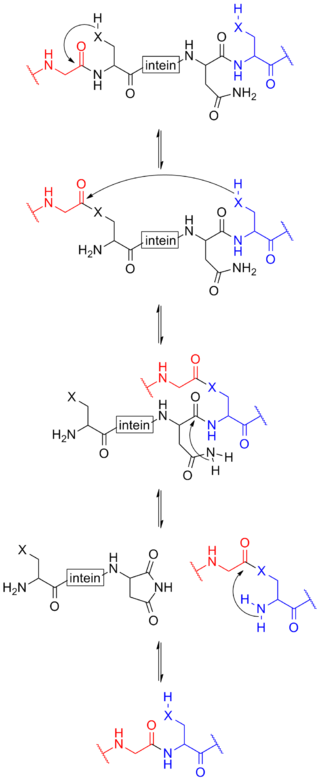
Protein splicing is an intramolecular reaction of a particular protein in which an internal protein segment is removed from a precursor protein with a ligation of C-terminal and N-terminal external proteins on both sides. The splicing junction of the precursor protein is mainly a cysteine or a serine, which are amino acids containing a nucleophilic side chain. The protein splicing reactions which are known now do not require exogenous cofactors or energy sources such as adenosine triphosphate (ATP) or guanosine triphosphate (GTP). Normally, splicing is associated only with pre-mRNA splicing. This precursor protein contains three segments—an N-extein followed by the intein followed by a C-extein. After splicing has taken place, the resulting protein contains the N-extein linked to the C-extein; this splicing product is also termed an extein.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.

The restriction endonuclease Fok1, naturally found in Flavobacterium okeanokoites, is a bacterial type IIS restriction endonuclease consisting of an N-terminal DNA-binding domain and a non sequence-specific DNA cleavage domain at the C-terminal. Once the protein is bound to duplex DNA via its DNA-binding domain at the 5'-GGATG-3' recognition site, the DNA cleavage domain is activated and cleaves the DNA at two locations, regardless of the nucleotide sequence at the cut site. The DNA is cut 9 nucleotides downstream of the motif on the forward strand, and 13 nucleotides downstream of the motif on the reverse strand, producing two sticky ends with 4-bp overhangs.
In molecular biology, a twintron is an intron-within-intron excised by sequential splicing reactions. A twintron is presumably formed by the insertion of a mobile intron into an existing intron.

I-CreI is a homing endonuclease whose gene was first discovered in the chloroplast genome of Chlamydomonas reinhardtii, a species of unicellular green algae. It is named for the facts that: it resides in an Intron; it was isolated from Clamydomonas reinhardtii; it was the first (I) such gene isolated from C. reinhardtii. Its gene resides in a group I intron in the 23S ribosomal RNA gene of the C. reinhardtii chloroplast, and I-CreI is only expressed when its mRNA is spliced from the primary transcript of the 23S gene. I-CreI enzyme, which functions as a homodimer, recognizes a 22-nucleotide sequence of duplex DNA and cleaves one phosphodiester bond on each strand at specific positions. I-CreI is a member of the LAGLIDADG family of homing endonucleases, all of which have a conserved LAGLIDADG amino acid motif that contributes to their associative domains and active sites. When the I-CreI-containing intron encounters a 23S allele lacking the intron, I-CreI enzyme "homes" in on the "intron-minus" allele of 23S and effects its parent intron's insertion into the intron-minus allele. Introns with this behavior are called mobile introns. Because I-CreI provides for its own propagation while conferring no benefit on its host, it is an example of selfish DNA.

EcoRV is a type II restriction endonuclease isolated from certain strains of Escherichia coli. It has the alternative name Eco32I.
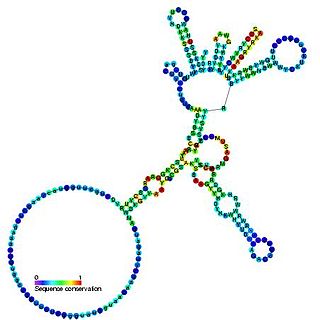
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

Nuclease S1 is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalyses the following chemical reaction

DNA-(apurinic or apyrimidinic site) lyase is an enzyme that in humans is encoded by the APEX1 gene.
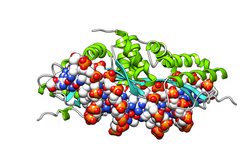
Restriction endonuclease (REase) EcoRII is an enzyme of restriction modification system (RM) naturally found in Escherichia coli, a Gram-negative bacteria. Its molecular mass is 45.2 kDa, being composed of 402 amino acids.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.
Meganucleases are endodeoxyribonucleases characterized by a large recognition site ; as a result this site generally occurs only once in any given genome. For example, the 18-base pair sequence recognized by the I-SceI meganuclease would on average require a genome twenty times the size of the human genome to be found once by chance. Meganucleases are therefore considered to be the most specific naturally occurring restriction enzymes.

Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases, and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ).

Cas9 is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.
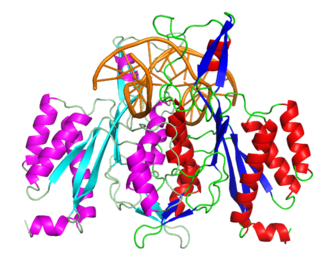
EcoRI is a restriction endonuclease enzyme isolated from species E. coli. It is a restriction enzyme that cleaves DNA double helices into fragments at specific sites, and is also a part of the restriction modification system. The Eco part of the enzyme's name originates from the species from which it was isolated - "E" denotes generic name which is "Escherichia" and "co" denotes species name, "coli" - while the R represents the particular strain, in this case RY13, and the I denotes that it was the first enzyme isolated from this strain.