
p53, also known as Tumor protein P53, cellular tumor antigen p53, or transformation-related protein 53 (TRP53) is a regulatory protein that is often mutated in human cancers. The p53 proteins are crucial in vertebrates, where they prevent cancer formation. As such, p53 has been described as "the guardian of the genome" because of its role in conserving stability by preventing genome mutation. Hence TP53 is classified as a tumor suppressor gene.

Ubiquitin is a small regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.

Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the DLD gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide.

Xanthine dehydrogenase, also known as XDH, is a protein that, in humans, is encoded by the XDH gene.

Nuclear factor erythroid 2-related factor 2 (NRF2), also known as nuclear factor erythroid-derived 2-like 2, is a transcription factor that in humans is encoded by the NFE2L2 gene. NRF2 is a basic leucine zipper (bZIP) protein that may regulate the expression of antioxidant proteins that protect against oxidative damage triggered by injury and inflammation, according to preliminary research. In vitro, NRF2 binds to antioxidant response elements (AREs) in the promoter regions of genes encoding cytoprotective proteins. NRF2 induces the expression of heme oxygenase 1 in vitro leading to an increase in phase II enzymes. NRF2 also inhibits the NLRP3 inflammasome.
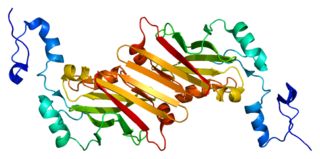
E3 ubiquitin-protein ligase SIAH1 is an enzyme that in humans is encoded by the SIAH1 gene.

In enzymology, a ribosyldihydronicotinamide dehydrogenase (quinone) (EC 1.10.99.2) is an enzyme that catalyzes the chemical reaction
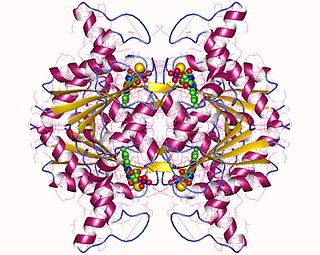
In enzymology, a NAD(P)H dehydrogenase (quinone) (EC 1.6.5.2) is an enzyme that catalyzes the chemical reaction

Methionine synthase reductase, also known as MSR, is an enzyme that in humans is encoded by the MTRR gene.
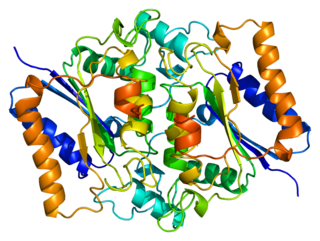
NAD(P)H dehydrogenase, quinone 2, also known as QR2, is a protein that in humans is encoded by the NQO2 gene. It is a phase II detoxification enzyme which can carry out two or four electron reductions of quinones. Its mechanism of reduction is through a ping-pong mechanism involving its FAD cofactor. Initially in a reductive phase NQO2 binds to reduced dihydronicotinamide riboside (NRH) electron donor, and mediates a hydride transfer from NRH to FAD. Then, in an oxidative phase, NQO2 binds to its quinone substrate and reduces the quinone to a dihydroquinone. Besides the two catalytic FAD, NQO2 also has two zinc ions. It is not clear whether the metal has a catalytic role. NQO2 is a paralog of NQO1.

Carbonyl reductase 1, also known as CBR1, is an enzyme which in humans is encoded by the CBR1 gene. The protein encoded by this gene belongs to the short-chain dehydrogenases/reductases (SDR) family, which function as NADPH-dependent oxidoreductases having wide specificity for carbonyl compounds, such as quinones, prostaglandins, and various xenobiotics. Alternatively spliced transcript variants have been found for this gene.

Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial is an enzyme that in humans is encoded by the ALDH4A1 gene.

Putative quinone oxidoreductase is an enzyme that in humans is encoded by the TP53I3 gene.

Apoptosis-inducing factor 2 (AIFM2), also known as ferroptosis suppressor protein 1 (FSP1), apoptosis-inducing factor-homologous mitochondrion-associated inducer of death (AMID), is a protein that in humans is encoded by the AIFM2 gene, also known as p53-responsive gene 3 (PRG3), on chromosome 10.

CYP27C1 is a protein that in humans is encoded by the CYP27C1 gene. The Enzyme Commission number (EC) for this protein is EC 1.14.19.53. The full accepted name is all-trans-retinol 3,4-desaturase and the EC number 1 classifies CYP27C1 as a oxidoreductase that acts on paired donor by reducing oxygen. It is also identifiable by the UniProt code Q4G0S4.

Renalase, FAD-dependent amine oxidase is an enzyme that in humans is encoded by the RNLS gene. Renalase is a flavin adenine dinucleotide-dependent amine oxidase that is secreted into the blood from the kidney.

NADH:ubiquinone reductase (non-electrogenic) (EC 1.6.5.9, NDH-2, ubiquinone reductase, coenzyme Q reductase, dihydronicotinamide adenine dinucleotide-coenzyme Q reductase, DPNH-coenzyme Q reductase, DPNH-ubiquinone reductase, NADH-coenzyme Q oxidoreductase, NADH-coenzyme Q reductase, NADH-CoQ oxidoreductase, NADH-CoQ reductase) is an enzyme with systematic name NADH:ubiquinone oxidoreductase. This enzyme catalyses the following chemical reaction:
Hydroquinone 1,2-dioxygenase (EC 1.13.11.66, hydroquinone dioxygenase) is an enzyme with systematic name benzene-1,4-diol:oxygen 1,2-oxidoreductase (decyclizing). This enzyme catalyses the following chemical reaction

WRAP53 is a gene implicated in cancer development. The name was coined in 2009 to describe the dual role of this gene, encoding both an antisense RNA that regulates the p53 tumor suppressor and a protein involved in DNA repair, telomere elongation and maintenance of nuclear organelles Cajal bodies.
Jasvinder K Gambhir is an Indian doctor, researcher and professor in the fields of clinical biochemistry, diabetology and cardiology. Dr Gambhir completed her master's in biochemistry in 1972 from Punjab University and PhD from Post Graduate Institute of Medical Education and Research She has an experience in the field of over 40 years. She is Senior professor and Head of Department, Biochemistry at University College of Medical Sciences, New Delhi and Senior Professor at School of Medical Sciences and Research Noida. Dr. Gambhir is also a member of American Association of Clinical Chemistry (AACC).
































