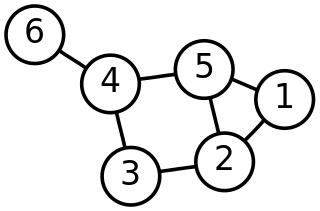
In mathematics, graph theory is the study of graphs, which are mathematical structures used to model pairwise relations between objects. A graph in this context is made up of vertices which are connected by edges. A distinction is made between undirected graphs, where edges link two vertices symmetrically, and directed graphs, where edges link two vertices asymmetrically. Graphs are one of the principal objects of study in discrete mathematics.
The Ising model, named after the physicists Ernst Ising and Wilhelm Lenz, is a mathematical model of ferromagnetism in statistical mechanics. The model consists of discrete variables that represent magnetic dipole moments of atomic "spins" that can be in one of two states. The spins are arranged in a graph, usually a lattice, allowing each spin to interact with its neighbors. Neighboring spins that agree have a lower energy than those that disagree; the system tends to the lowest energy but heat disturbs this tendency, thus creating the possibility of different structural phases. The model allows the identification of phase transitions as a simplified model of reality. The two-dimensional square-lattice Ising model is one of the simplest statistical models to show a phase transition.

In mathematics, random graph is the general term to refer to probability distributions over graphs. Random graphs may be described simply by a probability distribution, or by a random process which generates them. The theory of random graphs lies at the intersection between graph theory and probability theory. From a mathematical perspective, random graphs are used to answer questions about the properties of typical graphs. Its practical applications are found in all areas in which complex networks need to be modeled – many random graph models are thus known, mirroring the diverse types of complex networks encountered in different areas. In a mathematical context, random graph refers almost exclusively to the Erdős–Rényi random graph model. In other contexts, any graph model may be referred to as a random graph.
In the domain of physics and probability, a Markov random field (MRF), Markov network or undirected graphical model is a set of random variables having a Markov property described by an undirected graph. In other words, a random field is said to be a Markov random field if it satisfies Markov properties. The concept originates from the Sherrington–Kirkpatrick model.

In mathematics, and more specifically in graph theory, a multigraph is a graph which is permitted to have multiple edges, that is, edges that have the same end nodes. Thus two vertices may be connected by more than one edge.
In universal algebra and in model theory, a structure consists of a set along with a collection of finitary operations and relations that are defined on it.
In mathematics, computer science and digital electronics, a dependency graph is a directed graph representing dependencies of several objects towards each other. It is possible to derive an evaluation order or the absence of an evaluation order that respects the given dependencies from the dependency graph.
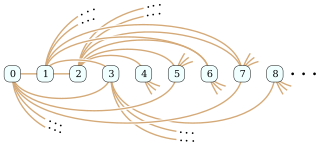
In the mathematical field of graph theory, the Rado graph, Erdős–Rényi graph, or random graph is a countably infinite graph that can be constructed by choosing independently at random for each pair of its vertices whether to connect the vertices by an edge. The names of this graph honor Richard Rado, Paul Erdős, and Alfréd Rényi, mathematicians who studied it in the early 1960s; it appears even earlier in the work of Wilhelm Ackermann. The Rado graph can also be constructed non-randomly, by symmetrizing the membership relation of the hereditarily finite sets, by applying the BIT predicate to the binary representations of the natural numbers, or as an infinite Paley graph that has edges connecting pairs of prime numbers congruent to 1 mod 4 that are quadratic residues modulo each other.

In the study of complex networks, a network is said to have community structure if the nodes of the network can be easily grouped into sets of nodes such that each set of nodes is densely connected internally. In the particular case of non-overlapping community finding, this implies that the network divides naturally into groups of nodes with dense connections internally and sparser connections between groups. But overlapping communities are also allowed. The more general definition is based on the principle that pairs of nodes are more likely to be connected if they are both members of the same community(ies), and less likely to be connected if they do not share communities. A related but different problem is community search, where the goal is to find a community that a certain vertex belongs to.

In the mathematical field of graph theory, the Erdős–Rényi model refers to one of two closely related models for generating random graphs or the evolution of a random network. These models are named after Hungarian mathematicians Paul Erdős and Alfréd Rényi, who introduced one of the models in 1959. Edgar Gilbert introduced the other model contemporaneously with and independently of Erdős and Rényi. In the model of Erdős and Rényi, all graphs on a fixed vertex set with a fixed number of edges are equally likely. In the model introduced by Gilbert, also called the Erdős–Rényi–Gilbert model, each edge has a fixed probability of being present or absent, independently of the other edges. These models can be used in the probabilistic method to prove the existence of graphs satisfying various properties, or to provide a rigorous definition of what it means for a property to hold for almost all graphs.
The constellation model is a probabilistic, generative model for category-level object recognition in computer vision. Like other part-based models, the constellation model attempts to represent an object class by a set of N parts under mutual geometric constraints. Because it considers the geometric relationship between different parts, the constellation model differs significantly from appearance-only, or "bag-of-words" representation models, which explicitly disregard the location of image features.

Modularity is a measure of the structure of networks or graphs which measures the strength of division of a network into modules. Networks with high modularity have dense connections between the nodes within modules but sparse connections between nodes in different modules. Modularity is often used in optimization methods for detecting community structure in networks. Biological networks, including animal brains, exhibit a high degree of modularity. However, modularity maximization is not statistically consistent, and finds communities in its own null model, i.e. fully random graphs, and therefore it cannot be used to find statistically significant community structures in empirical networks. Furthermore, it has been shown that modularity suffers a resolution limit and, therefore, it is unable to detect small communities.
In computer science, lexicographic breadth-first search or Lex-BFS is a linear time algorithm for ordering the vertices of a graph. The algorithm is different from a breadth-first search, but it produces an ordering that is consistent with breadth-first search.
Graphical models have become powerful frameworks for protein structure prediction, protein–protein interaction, and free energy calculations for protein structures. Using a graphical model to represent the protein structure allows the solution of many problems including secondary structure prediction, protein-protein interactions, protein-drug interaction, and free energy calculations.

The stochastic block model is a generative model for random graphs. This model tends to produce graphs containing communities, subsets of nodes characterized by being connected with one another with particular edge densities. For example, edges may be more common within communities than between communities. Its mathematical formulation was first introduced in 1983 in the field of social network analysis by Paul W. Holland et al. The stochastic block model is important in statistics, machine learning, and network science, where it serves as a useful benchmark for the task of recovering community structure in graph data.

In network science, the configuration model is a method for generating random networks from a given degree sequence. It is widely used as a reference model for real-life social networks, because it allows the modeler to incorporate arbitrary degree distributions.

Maximum-entropy random graph models are random graph models used to study complex networks subject to the principle of maximum entropy under a set of structural constraints, which may be global, distributional, or local.

In applied mathematics, the soft configuration model (SCM) is a random graph model subject to the principle of maximum entropy under constraints on the expectation of the degree sequence of sampled graphs. Whereas the configuration model (CM) uniformly samples random graphs of a specific degree sequence, the SCM only retains the specified degree sequence on average over all network realizations; in this sense the SCM has very relaxed constraints relative to those of the CM ("soft" rather than "sharp" constraints). The SCM for graphs of size has a nonzero probability of sampling any graph of size , whereas the CM is restricted to only graphs having precisely the prescribed connectivity structure.
In statistical mechanics, probability theory, graph theory, etc. the random cluster model is a random graph that generalizes and unifies the Ising model, Potts model, and percolation model. It is used to study random combinatorial structures, electrical networks, etc. It is also referred to as the RC model or sometimes the FK representation after its founders Cees Fortuin and Piet Kasteleyn.
Flag algebras are an important computational tool in the field of graph theory which have a wide range of applications in homomorphism density and related topics. Roughly, they formalize the notion of adding and multiplying homomorphism densities and set up a framework to solve graph homomorphism inequalities with computers by reducing them to semidefinite programming problems. Originally introduced by Alexander Razborov in a 2007 paper, the method has since come to solve numerous difficult, previously unresolved graph theoretic questions. These include the question regarding the region of feasible edge density, triangle density pairs and the maximum number of pentagons in triangle free graphs.












