
Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
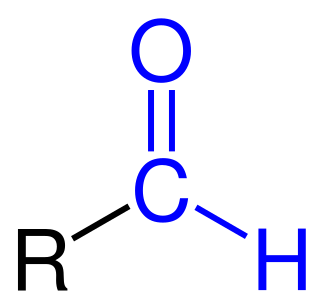
Formylation refers to any chemical processes in which a compound is functionalized with a formyl group (-CH=O). In organic chemistry, the term is most commonly used with regards to aromatic compounds. In biochemistry the reaction is catalysed by enzymes such as formyltransferases.
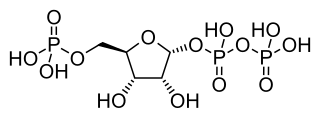
Phosphoribosyl pyrophosphate (PRPP) is a pentose phosphate. It is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, as well as in pyrimidine nucleotide formation. Hence it is a building block for DNA and RNA. The vitamins thiamine and cobalamin, and the amino acid tryptophan also contain fragments derived from PRPP. It is formed from ribose 5-phosphate (R5P) by the enzyme ribose-phosphate diphosphokinase:

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).
Phosphoribosylformylglycinamidine cyclo-ligase is the fifth enzyme in the de novo synthesis of purine nucleotides. It catalyzes the reaction to form 5-aminoimidazole ribotide (AIR) from formylglycinamidine-ribonucleotide FGAM. This reaction closes the ring and produces a 5-membered imidazole ring of the purine nucleus (AIR):
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.
The enzyme Phosphoribosylaminoimidazole carboxylase, or AIR carboxylase (EC 4.1.1.21) is involved in nucleotide biosynthesis and in particular in purine biosynthesis. It catalyzes the conversion of 5'-phosphoribosyl-5-aminoimidazole ("AIR") into 5'-phosphoribosyl-4-carboxy-5-aminoimidazole ("CAIR") as described in the reaction:
In enzymology, a 5-(carboxyamino)imidazole ribonucleotide mutase is an enzyme that catalyzes the chemical reaction

In enzymology, a phosphoribosylanthranilate isomerase (PRAI) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.
In enzymology, a 5-(carboxyamino)imidazole ribonucleotide synthase (EC 6.3.4.18) is an enzyme that catalyzes the chemical reaction
Phosphoribosylamine—glycine ligase, also known as glycinamide ribonucleotide synthetase (GARS), (EC 6.3.4.13) is an enzyme that catalyzes the chemical reaction

In enzymology, a phosphoribosylformylglycinamidine synthase (EC 6.3.5.3) is an enzyme that catalyzes the chemical reaction
In enzymology, an IMP cyclohydrolase (EC 3.5.4.10) is an enzyme that catalyzes the chemical reaction

In enzymology, a nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

5′-Phosphoribosyl-5-aminoimidazole is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from AIR. It is an intermediate in the adenine pathway and is synthesized from 5′-phosphoribosylformylglycinamidine by AIR synthetase.

Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2, 2-amino-N-ribosylacetamide 5'-phosphate transformylase, GAR formyltransferase, GAR transformylase, glycinamide ribonucleotide transformylase, GAR TFase, 5,10-methenyltetrahydrofolate:2-amino-N-ribosylacetamide ribonucleotide transformylase) is an enzyme with systematic name 10-formyltetrahydrofolate:5'-phosphoribosylglycinamide N-formyltransferase. This enzyme catalyses the following chemical reaction
Phosphomethylpyrimidine synthase is an enzyme with systematic name 5-amino-1-(5-phospho-D-ribosyl)imidazole formate-lyase . This enzyme catalyses the following chemical reaction

















