Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.

Umami, or savoriness, is one of the five basic tastes. It has been described as savory and is characteristic of broths and cooked meats.

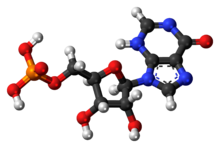
Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine. It is an ester of phosphoric acid and the nucleoside adenosine. As a substituent it takes the form of the prefix adenylyl-.

Disodium inosinate (E631) is the disodium salt of inosinic acid with the chemical formula C10H11N4Na2O8P. It is used as a food additive and often found in instant noodles, potato chips, and a variety of other snacks.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.

Disodium guanylate, also known as sodium 5'-guanylate and disodium 5'-guanylate, is a natural sodium salt of the flavor enhancing nucleotide guanosine monophosphate (GMP). Disodium guanylate is a food additive with the E number E627. It is commonly used in conjunction with glutamic acid.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
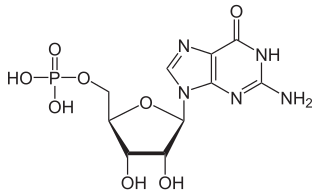
Guanosine monophosphate (GMP), also known as 5′-guanidylic acid or guanylic acid, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid with the nucleoside guanosine. GMP consists of the phosphate group, the pentose sugar ribose, and the nucleobase guanine; hence it is a ribonucleoside monophosphate. Guanosine monophosphate is commercially produced by microbial fermentation.

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

Ribose 5-phosphate (R5P) is both a product and an intermediate of the pentose phosphate pathway. The last step of the oxidative reactions in the pentose phosphate pathway is the production of ribulose 5-phosphate. Depending on the body's state, ribulose 5-phosphate can reversibly isomerize to ribose 5-phosphate. Ribulose 5-phosphate can alternatively undergo a series of isomerizations as well as transaldolations and transketolations that result in the production of other pentose phosphates as well as fructose 6-phosphate and glyceraldehyde 3-phosphate.
Phosphoribosylformylglycinamidine cyclo-ligase is the fifth enzyme in the de novo synthesis of purine nucleotides. It catalyzes the reaction to form 5-aminoimidazole ribotide (AIR) from formylglycinamidine-ribonucleotide FGAM. This reaction closes the ring and produces a 5-membered imidazole ring of the purine nucleus (AIR):
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.

Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme responsible for catalyzing the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) into 5-phosphoribosyl-1-amine (PRA), using the amine group from a glutamine side-chain. This is the committing step in de novo purine synthesis. In humans it is encoded by the PPAT gene. ATase is a member of the purine/pyrimidine phosphoribosyltransferase family.
Ribose-phosphate diphosphokinase is an enzyme that converts ribose 5-phosphate into phosphoribosyl pyrophosphate (PRPP). It is classified under EC 2.7.6.1.

5′-Phosphoribosyl-5-aminoimidazole is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from AIR. It is an intermediate in the adenine pathway and is synthesized from 5′-phosphoribosylformylglycinamidine by AIR synthetase.
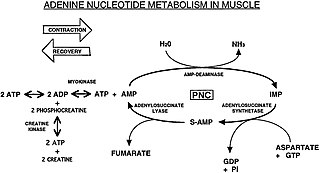
The Purine Nucleotide Cycle is a metabolic pathway in protein metabolism requiring the amino acids aspartate and glutamate. The cycle is used to regulate the levels of adenine nucleotides, in which ammonia and fumarate are generated. AMP converts into IMP and the byproduct ammonia. IMP converts to S-AMP (adenylosuccinate), which then converts to AMP and the byproduct fumarate. The fumarate goes on to produce ATP (energy) via oxidative phosphorylation as it enters the Krebs cycle and then the electron transport chain. Lowenstein first described this pathway and outlined its importance in processes including amino acid catabolism and regulation of flux through glycolysis and the Krebs cycle.
The gua operon is responsible for regulating the synthesis of guanosine mono phosphate (GMP), a purine nucleotide, from inosine monophosphate. It consists of two structural genes guaB (encodes for IMP dehydrogenase or and guaA apart from the promoter and operator region.
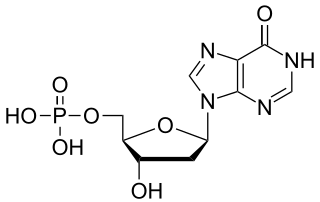
Deoxyinosine monophosphate (dIMP) is a nucleoside monophosphate and a derivative of inosinic acid. It can be formed by the deamination of the purine base in deoxyadenosine monophosphate (dAMP). The enzyme deoxyribonucleoside triphosphate pyrophosphohydrolase, encoded by YJR069C in S. cerevisiae and containing (d)ITPase and (d)XTPase activities, hydrolyses dITP, resulting in the release of pyrophosphate and dIMP.