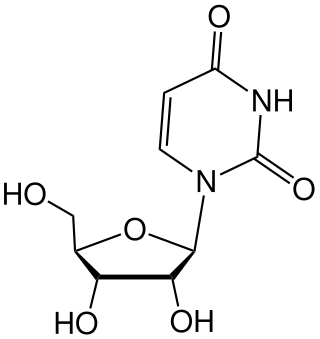
Uridine (symbol U or Urd) is a glycosylated pyrimidine analog containing uracil attached to a ribose ring (or more specifically, a ribofuranose) via a β-N1-glycosidic bond. The analog is one of the five standard nucleosides which make up nucleic acids, the others being adenosine, thymidine, cytidine and guanosine. The five nucleosides are commonly abbreviated to their symbols, U, A, dT, C, and G, respectively. However, thymidine is more commonly written as 'dT' ('d' represents 'deoxy') as it contains a 2'-deoxyribofuranose moiety rather than the ribofuranose ring found in uridine. This is because thymidine is found in deoxyribonucleic acid (DNA) and usually not in ribonucleic acid (RNA). Conversely, uridine is found in RNA and not DNA. The remaining three nucleosides may be found in both RNA and DNA. In RNA, they would be represented as A, C and G whereas in DNA they would be represented as dA, dC and dG.

Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbone attached to adenine and two phosphate groups bonded to the 5 carbon atom of ribose. The diphosphate group of ADP is attached to the 5’ carbon of the sugar backbone, while the adenine attaches to the 1’ carbon.

Anabolism is the set of metabolic pathways that construct macromolecules like DNA or RNA from smaller units. These reactions require energy, known also as an endergonic process. Anabolism is the building-up aspect of metabolism, whereas catabolism is the breaking-down aspect. Anabolism is usually synonymous with biosynthesis.
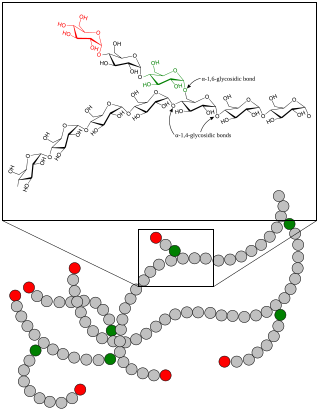
Glycogenolysis is the breakdown of glycogen (n) to glucose-1-phosphate and glycogen (n-1). Glycogen branches are catabolized by the sequential removal of glucose monomers via phosphorolysis, by the enzyme glycogen phosphorylase.
Carbohydrate metabolism is the whole of the biochemical processes responsible for the metabolic formation, breakdown, and interconversion of carbohydrates in living organisms.
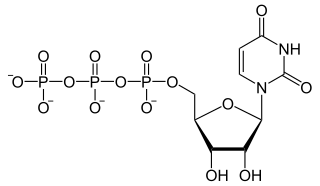
Uridine-5′-triphosphate (UTP) is a pyrimidine nucleoside triphosphate, consisting of the organic base uracil linked to the 1′ carbon of the ribose sugar, and esterified with tri-phosphoric acid at the 5′ position. Its main role is as substrate for the synthesis of RNA during transcription. UTP is the precursor for the production of CTP via CTP synthetase. UTP can be biosynthesized from UDP by Nucleoside Diphosphate Kinase after using the phosphate group from ATP. UDP + ATP ⇌ UTP + ADP; both UTP and ATP are energetically equal.

Glucose 6-phosphate is a glucose sugar phosphorylated at the hydroxy group on carbon 6. This dianion is very common in cells as the majority of glucose entering a cell will become phosphorylated in this way.
In organic chemistry, a tetrose is a monosaccharide with 4 carbon atoms. They have either an aldehyde functional group in position 1 (aldotetroses) or a ketone group in position 2 (ketotetroses).
Bioenergetics is a field in biochemistry and cell biology that concerns energy flow through living systems. This is an active area of biological research that includes the study of the transformation of energy in living organisms and the study of thousands of different cellular processes such as cellular respiration and the many other metabolic and enzymatic processes that lead to production and utilization of energy in forms such as adenosine triphosphate (ATP) molecules. That is, the goal of bioenergetics is to describe how living organisms acquire and transform energy in order to perform biological work. The study of metabolic pathways is thus essential to bioenergetics.

Galactosamine is a hexosamine derived from galactose with the molecular formula C6H13NO5. This amino sugar is a constituent of some glycoprotein hormones such as follicle-stimulating hormone (FSH) and luteinizing hormone (LH).

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

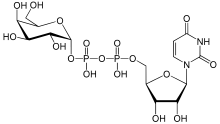
Uridine diphosphate glucose is a nucleotide sugar. It is involved in glycosyltransferase reactions in metabolism.

D-Xylulose 5-phosphate (D-xylulose-5-P) is an intermediate in the pentose phosphate pathway. It is a ketose sugar formed from ribulose-5-phosphate by ribulose-5-phosphate epimerase. In the non-oxidative branch of the pentose phosphate pathway, xylulose-5-phosphate acts as a donor of two-carbon ketone groups in transketolase reactions.
UTP—glucose-1-phosphate uridylyltransferase also known as glucose-1-phosphate uridylyltransferase is an enzyme involved in carbohydrate metabolism. It synthesizes UDP-glucose from glucose-1-phosphate and UTP; i.e.,

D-Galactose-1-phosphate is an intermediate in the intraconversion of glucose and uridine diphosphate galactose. It is formed from galactose by galactokinase.The improper metabolism of galactose-1-phosphate is a characteristic of galactosemia. The Leloir pathway is responsible for such metabolism of galactose and its intermediate, galactose-1-phosphate. Deficiency of enzymes found in this pathway can result in galactosemia; therefore, diagnosis of this genetic disorder occasionally involves measuring the concentration of these enzymes. One of such enzymes is galactose-1-phosphate uridylyltransferase (GALT). The enzyme catalyzes the transfer of a UDP-activator group from UDP-glucose to galactose-1-phosphate. Although the cause of enzyme deficiency in the Leloir pathway is still disputed amongst researchers, some studies suggest that protein misfolding of GALT, which may lead to an unfavorable conformational change that impacts its thermal stability and substrate-binding affinity, may play a role in the deficiency of GALT in Type 1 galactosemia. Increase in galactitol concentration can be seen in patients with galactosemia; putting patients at higher risk for presenile cataract.

Deoxyuridine monophosphate (dUMP), also known as deoxyuridylic acid or deoxyuridylate in its conjugate acid and conjugate base forms, respectively, is a deoxynucleotide.

The enzyme UDP-glucose 4-epimerase, also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.
In enzymology, an UTP—hexose-1-phosphate uridylyltransferase is an enzyme that catalyzes the chemical reaction

Inborn errors of carbohydrate metabolism are inborn error of metabolism that affect the catabolism and anabolism of carbohydrates.
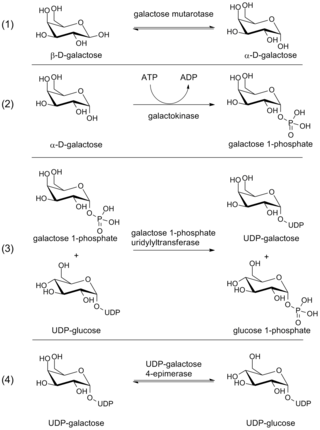
The Leloir pathway is a metabolic pathway for the catabolism of D-galactose. It is named after Luis Federico Leloir, who first described it.