
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.

Ubiquitin is a small regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.

A ubiquitin ligase is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases regulates diverse areas such as cell trafficking, DNA repair, and signaling and is of profound importance in cell biology. E3 ligases are also key players in cell cycle control, mediating the degradation of cyclins, as well as cyclin dependent kinase inhibitor proteins. The human genome encodes over 600 putative E3 ligases, allowing for tremendous diversity in substrates.
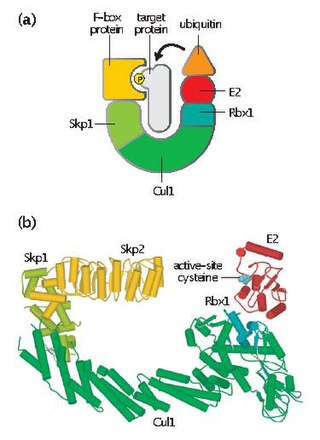
Skp, Cullin, F-box containing complex is a multi-protein E3 ubiquitin ligase complex that catalyzes the ubiquitination of proteins destined for 26S proteasomal degradation. Along with the anaphase-promoting complex, SCF has important roles in the ubiquitination of proteins involved in the cell cycle. The SCF complex also marks various other cellular proteins for destruction.

Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which can target a protein for degradation via a proteasome. This covalent bond of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms. Many processes such as cell division, immune responses and embryonic development are also regulated by post-translational modification by ubiquitin and ubiquitin-like proteins.
Ubiquitin-conjugating enzymes, also known as E2 enzymes and more rarely as ubiquitin-carrier enzymes, perform the second step in the ubiquitination reaction that targets a protein for degradation via the proteasome. The ubiquitination process covalently attaches ubiquitin, a short protein of 76 amino acids, to a lysine residue on the target protein. Once a protein has been tagged with one ubiquitin molecule, additional rounds of ubiquitination form a polyubiquitin chain that is recognized by the proteasome's 19S regulatory particle, triggering the ATP-dependent unfolding of the target protein that allows passage into the proteasome's 20S core particle, where proteases degrade the target into short peptide fragments for recycling by the cell.

S-phase kinase-associated protein 2 is an enzyme that in humans is encoded by the SKP2 gene.

Cullin-4A is a protein that in humans is encoded by the CUL4A gene. CUL4A belongs to the cullin family of ubiquitin ligase proteins and is highly homologous to the CUL4B protein. CUL4A regulates numerous key processes such as DNA repair, chromatin remodeling, spermatogenesis, haematopoiesis and the mitotic cell cycle. As a result, CUL4A has been implicated in several cancers and the pathogenesis of certain viruses including HIV. A component of a CUL4A complex, Cereblon, was discovered to be a major target of the teratogenic agent thalidomide.

Ubiquitin D is a protein that in humans is encoded by the UBD gene, also known as FAT10. UBD acts like ubiquitin, by covalently modifying proteins and tagging them for destruction in the proteasome.

Cullin-4B is a protein that in humans is encoded by the CUL4B gene which is located on the X chromosome. CUL4B has high sequence similarity with CUL4A, with which it shares certain E3 ubiquitin ligase functions. CUL4B is largely expressed in the nucleus and regulates several key functions including: cell cycle progression, chromatin remodeling and neurological and placental development in mice. In humans, CUL4B has been implicated in X-linked intellectual disability and is frequently mutated in pancreatic adenocarcinomas and a small percentage of various lung cancers. Viruses such as HIV can also co-opt CUL4B-based complexes to promote viral pathogenesis. CUL4B complexes containing Cereblon are also targeted by the teratogenic drug thalidomide.

Cullin 3 is a protein that in humans is encoded by the CUL3 gene.

26S proteasome non-ATPase regulatory subunit 14, also known as 26S proteasome non-ATPase subunit Rpn11, is an enzyme that in humans is encoded by the PSMD14 gene. This protein is one of the 19 essential subunits of the complete assembled 19S proteasome complex. Nine subunits Rpn3, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn11, SEM1, and Rpn12 form the lid sub complex of the 19S regulatory particle of the proteasome complex.

Cereblon is a protein that in humans is encoded by the CRBN gene. The gene that encodes the cereblon protein is found on the human chromosome 3, on the short arm at position p26.3 from base pair 3,190,676 to base pair 3,221,394. CRBN orthologs are highly conserved from plants to humans.

Cdc4 is a substrate recognition component of the SCF ubiquitin ligase complex, which acts as a mediator of ubiquitin transfer to target proteins, leading to their subsequent degradation via the ubiquitin-proteasome pathway. Cdc4 targets primarily cell cycle regulators for proteolysis. It serves the function of an adaptor that brings target molecules to the core SCF complex. Cdc4 was originally identified in the model organism Saccharomyces cerevisiae. CDC4 gene function is required at G1/S and G2/M transitions during mitosis and at various stages during meiosis.
Craig M. Crews is an American scientist at Yale University known for his contributions to chemical biology. He is known for his contributions to the field of induced proximity through his work in creating heterobifunctional molecules that "hijack" cellular processes by inducing the interaction of two proteins inside a living cell. His initial work focused on the discovery of PROteolysis-TArgeting Chimeras (PROTACs) to trigger degradation of disease-causing proteins, a process known as targeted protein degradation (TPD), and he has since developed new versions of -TACs to leverage other cellular processes and protein families to treat disease.
Raymond Joseph Deshaies is an American biochemist and cell biologist. He is senior vice president of global research at Amgen and a visiting associate at the California Institute of Technology (Caltech). Prior to that, he was a professor of biology at Caltech and an investigator of the Howard Hughes Medical Institute. He is also the co-founder of the biotechnology companies Proteolix and Cleave Biosciences. His research focuses on mechanisms and regulation of protein homeostasis in eukaryotic cells, with a particular focus on how proteins are conjugated with ubiquitin and degraded by the proteasome.

Daniel K. Nomura is an American chemical biologist and Professor of Chemical Biology and Molecular Therapeutics at the University of California, Berkeley, in the Departments of Chemistry and Molecular & Cell Biology. His work employs chemoproteomic approaches to develop small molecule therapeutics and therapeutic modalities against traditionally "undruggable" proteins.
Alessio Ciulli is an Italian British biochemist. Currently, he is the Professor of Chemical & Structural Biology at the School of Life Sciences, University of Dundee, where he founded and directs Dundee' new Centre for Targeted Protein Degradation (CeTPD). He is also the scientific co-founder and advisor of Amphista Therapeutics.
Molecular glue refers to a class of chemical compounds or molecules that play a crucial role in binding and stabilizing protein-protein interactions in biological systems. These molecules act as "glue" by enhancing the affinity between proteins, ultimately influencing various cellular processes. Molecular glue compounds have gained significant attention in the fields of drug discovery, chemical biology, and fundamental research due to their potential to modulate protein interactions, and thus, impact various cellular pathways. They have unlocked avenues in medicine previously thought to be "undruggable".
Chimeric small molecule therapeutics are a class of drugs designed with multiple active domains to operate outside of the typical protein inhibition model. While most small molecule drugs inhibit target proteins by binding their active site, chimerics form protein-protein ternary structures to induce degradation or, less frequently, other protein modifications.















