
Protein biosynthesis is a core biological process, occurring inside cells, balancing the loss of cellular proteins through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.
In genetics, a transcription terminator is a section of nucleic acid sequence that marks the end of a gene or operon in genomic DNA during transcription. This sequence mediates transcriptional termination by providing signals in the newly synthesized transcript RNA that trigger processes which release the transcript RNA from the transcriptional complex. These processes include the direct interaction of the mRNA secondary structure with the complex and/or the indirect activities of recruited termination factors. Release of the transcriptional complex frees RNA polymerase and related transcriptional machinery to begin transcription of new mRNAs.
This is a list of topics in molecular biology. See also index of biochemistry articles.
RNA polymerase 1 is, in higher eukaryotes, the polymerase that only transcribes ribosomal RNA, a type of RNA that accounts for over 50% of the total RNA synthesized in a cell.
In molecular biology, a termination factor is a protein that mediates the termination of RNA transcription by recognizing a transcription terminator and causing the release of the newly made mRNA. This is part of the process that regulates the transcription of RNA to preserve gene expression integrity and are present in both eukaryotes and prokaryotes, although the process in bacteria is more widely understood. The most extensively studied and detailed transcriptional termination factor is the Rho (ρ) protein of E. coli.
In eukaryote cells, RNA polymerase III is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA and other small RNAs.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.
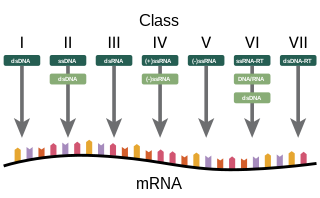
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.

Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end, which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end, which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information.
Antitermination is the prokaryotic cell's aid to fix premature termination of RNA synthesis during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.

Intrinsic, or rho-independent termination, is a process in prokaryotes to signal the end of transcription and release the newly constructed RNA molecule. In prokaryotes such as E. coli, transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a Stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil and adenine are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly-uracil sequence. The weak adenine-uracil bonds lower the energy of destabilization for the RNA-DNA duplex, allowing it to unwind and dissociate from the RNA polymerase. Overall, the modified RNA structure is what terminates transcription.
Post-transcriptional regulation is the control of gene expression at the RNA level. It occurs once the RNA polymerase has been attached to the gene's promoter and is synthesizing the nucleotide sequence. Therefore, as the name indicates, it occurs between the transcription phase and the translation phase of gene expression. These controls are critical for the regulation of many genes across human tissues. It also plays a big role in cell physiology, being implicated in pathologies such as cancer and neurodegenerative diseases.
Translational regulation refers to the control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms.
Transcription-translation coupling is a mechanism of gene expression regulation in which synthesis of an mRNA (transcription) is affected by its concurrent decoding (translation). In prokaryotes, mRNAs are translated while they are transcribed. This allows communication between RNA polymerase, the multisubunit enzyme that catalyzes transcription, and the ribosome, which catalyzes translation. Coupling involves both direct physical interactions between RNA polymerase and the ribosome, as well as ribosome-induced changes to the structure and accessibility of the intervening mRNA that affect transcription.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology, cell biology, and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.







