
Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.
The peptidyl transferase is an aminoacyltransferase as well as the primary enzymatic function of the ribosome, which forms peptide bonds between adjacent amino acids using tRNAs during the translation process of protein biosynthesis. The substrates for the peptidyl transferase reaction are two tRNA molecules, one bearing the growing peptide chain and the other bearing the amino acid that will be added to the chain. The peptidyl chain and the amino acids are attached to their respective tRNAs via ester bonds to the O atom at the CCA-3' ends of these tRNAs. Peptidyl transferase is an enzyme that catalyzes the addition of an amino acid residue in order to grow the polypeptide chain in protein synthesis. It is located in the large ribosomal subunit, where it catalyzes the peptide bond formation. It is composed entirely of RNA. The alignment between the CCA ends of the ribosome-bound peptidyl tRNA and aminoacyl tRNA in the peptidyl transferase center contribute to its ability to catalyze these reactions. This reaction occurs via nucleophilic displacement. The amino group of the aminoacyl tRNA attacks the terminal carboxyl group of the peptidyl tRNA. Peptidyl transferase activity is carried out by the ribosome. Peptidyl transferase activity is not mediated by any ribosomal proteins but by ribosomal RNA (rRNA), a ribozyme. Ribozymes are the only enzymes which are not made up of proteins, but ribonucleotides. All other enzymes are made up of proteins. This RNA relic is the most significant piece of evidence supporting the RNA World hypothesis.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.

Lincosamides are a class of antibiotics, which include lincomycin, clindamycin, and pirlimycin.

The 5S ribosomal RNA is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life, with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S).

50S is the larger subunit of the 70S ribosome of prokaryotes, i.e. bacteria and archaea. It is the site of inhibition for antibiotics such as macrolides, chloramphenicol, clindamycin, and the pleuromutilins. It includes the 5S ribosomal RNA and 23S ribosomal RNA.

The prokaryotic small ribosomal subunit, or 30S subunit, is the smaller subunit of the 70S ribosome found in prokaryotes. It is a complex of the 16S ribosomal RNA (rRNA) and 19 proteins. This complex is implicated in the binding of transfer RNA to messenger RNA (mRNA). The small subunit is responsible for the binding and the reading of the mRNA during translation. The small subunit, both the rRNA and its proteins, complexes with the large 50S subunit to form the 70S prokaryotic ribosome in prokaryotic cells. This 70S ribosome is then used to translate mRNA into proteins.
Ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. The 60S subunit is the large subunit of eukaryotic 80S ribosomes. It is structurally and functionally related to the 50S subunit of 70S prokaryotic ribosomes. However, the 60S subunit is much larger than the prokaryotic 50S subunit and contains many additional protein segments, as well as ribosomal RNA expansion segments.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.

A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins.
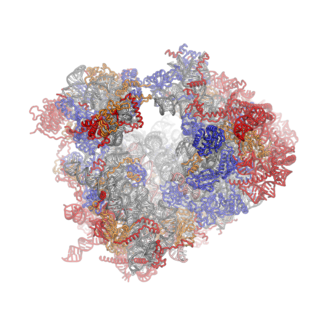
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes are much larger than prokaryotic ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain dozens of ribosomal proteins arranged on a scaffold composed of ribosomal RNA (rRNA). The small subunit monitors the complementarity between tRNA anticodon and mRNA, while the large subunit catalyzes peptide bond formation.

Kasugamycin (Ksg) is an aminoglycoside antibiotic that was originally isolated in 1965, from Streptomyces kasugaensis, a Streptomyces strain found near the Kasuga shrine in Nara, Japan. Kasugamycin was discovered by Hamao Umezawa, who also discovered kanamycin and bleomycin, as a drug that prevent growth of a fungus causing rice blast disease. It was later found to inhibit bacterial growth also. It exists as a white, crystalline substance with the chemical formula C14H28ClN3O10 (kasugamycin hydrochloride). It is also known as kasumin.
The P-site is the second binding site for tRNA in the ribosome. The other two sites are the A-site (aminoacyl), which is the first binding site in the ribosome, and the E-site (exit), the third. During protein translation, the P-site holds the tRNA which is linked to the growing polypeptide chain. When a stop codon is reached, the peptidyl-tRNA bond of the tRNA located in the P-site is cleaved releasing the newly synthesized protein. During the translocation step of the elongation phase, the mRNA is advanced by one codon, coupled to movement of the tRNAs from the ribosomal A to P and P to E sites, catalyzed by elongation factor EF-G.

In molecular biology, the CRM domain is an approximately 100-amino acid RNA-binding domain. The name CRM has been suggested to reflect the functions established for four characterised members of the family: Zea mays (Maize) CRS1, CAF1 and CAF2 proteins and the Escherichia coli protein YhbY. Proteins containing the CRM domain are found in eubacteria, archaea, and plants. The CRM domain is represented as a stand-alone protein in archaea and bacteria, and in single- and multi-domain proteins in plants. It has been suggested that prokaryotic CRM proteins existed as ribosome-associated proteins prior to the divergence of archaea and bacteria, and that they were co-opted in the plant lineage as RNA binding modules by incorporation into diverse protein contexts. Plant CRM domains are predicted to reside not only in the chloroplast, but also in the mitochondrion and the nucleo/cytoplasmic compartment. The diversity of the CRM domain family in plants suggests a diverse set of RNA targets.

EF-P is an essential protein that in bacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.

Helix 69 is a hairpin RNA structure containing 19 nucleotides in large subunit of the ribosome. Ribosome consists of large and small subunits joined with inter subunit bridges. Helix 69 interacts with the helix 44 (h44) of the small subunit to form the largest interface of two subunits called inter-subunit bridge B2a, one of the most conserved regions of the ribosome. Helix 69 is proposed to be a good drug target for antibacterial drugs. Many of the recent crystal structures have shown the involvement of this hairpin in different stages of the protein translation process. By targeting bacterial helix 69 specifically, protein synthesis in bacteria could be halted thus killing the bacteria.
In molecular biology, VAR1 protein domain, otherwise known as variant protein 1, is a ribosomal protein that forms part of the small ribosomal subunit in yeast mitochondria. Mitochondria possess their own ribosomes responsible for the synthesis of a small number of proteins encoded by the mitochondrial genome. VAR1 is the only protein in the yeast mitochondrial ribosome to be encoded in the mitochondria - the remaining approximately 80 ribosomal proteins are encoded in the nucleus. VAR1 along with 15S rRNA are necessary for the formation of mature 37S subunits.
Ribosomal L28e protein family is a family of evolutionarily related proteins. Members include 60S ribosomal protein L28.















