
Molybdenum is a chemical element; it has symbol Mo and atomic number 42. The name derived from Ancient Greek Μόλυβδος molybdos, meaning lead, since its ores were confused with lead ores. Molybdenum minerals have been known throughout history, but the element was discovered in 1778 by Carl Wilhelm Scheele. The metal was first isolated in 1781 by Peter Jacob Hjelm.
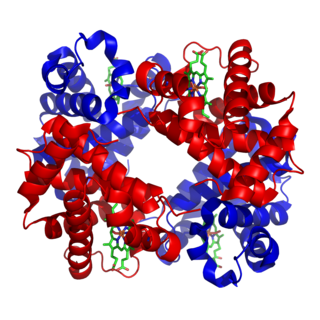
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins contain zinc-binding protein domains although there may be up to 3000 human zinc metalloproteins.
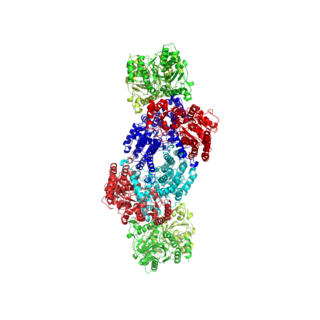
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.

Aconitase is an enzyme that catalyses the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle, a non-redox-active process.

Fumarase is an enzyme that catalyzes the reversible hydration/dehydration of fumarate to malate. Fumarase comes in two forms: mitochondrial and cytosolic. The mitochondrial isoenzyme is involved in the Krebs cycle and the cytosolic isoenzyme is involved in the metabolism of amino acids and fumarate. Subcellular localization is established by the presence of a signal sequence on the amino terminus in the mitochondrial form, while subcellular localization in the cytosolic form is established by the absence of the signal sequence found in the mitochondrial variety.
DMSO reductase is a molybdenum-containing enzyme that catalyzes reduction of dimethyl sulfoxide (DMSO) to dimethyl sulfide (DMS). This enzyme serves as the terminal reductase under anaerobic conditions in some bacteria, with DMSO being the terminal electron acceptor. During the course of the reaction, the oxygen atom in DMSO is transferred to molybdenum, and then reduced to water.

Molybdopterins are a class of cofactors found in most molybdenum-containing and all tungsten-containing enzymes. Synonyms for molybdopterin are: MPT and pyranopterin-dithiolate. The nomenclature for this biomolecule can be confusing: Molybdopterin itself contains no molybdenum; rather, this is the name of the ligand that will bind the active metal. After molybdopterin is eventually complexed with molybdenum, the complete ligand is usually called molybdenum cofactor.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
Nitrile hydratases are mononuclear iron or non-corrinoid cobalt enzymes that catalyse the hydration of diverse nitriles to their corresponding amides
In chemistry, a (redox) non-innocent ligand is a ligand in a metal complex where the oxidation state is not clear. Typically, complexes containing non-innocent ligands are redox active at mild potentials. The concept assumes that redox reactions in metal complexes are either metal or ligand localized, which is a simplification, albeit a useful one.
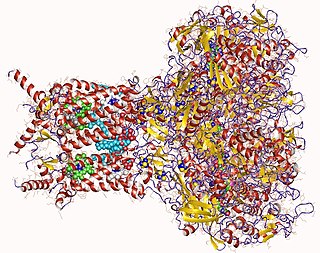
Formate dehydrogenases are a set of enzymes that catalyse the oxidation of formate to carbon dioxide, donating the electrons to a second substrate, such as NAD+ in formate:NAD+ oxidoreductase (EC 1.17.1.9) or to a cytochrome in formate:ferricytochrome-b1 oxidoreductase (EC 1.2.2.1). This family of enzymes has attracted attention as inspiration or guidance on methods for the carbon dioxide fixation, relevant to global warming.
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction
A transition metal oxo complex is a coordination complex containing an oxo ligand. Formally O2-, an oxo ligand can be bound to one or more metal centers, i.e. it can exist as a terminal or (most commonly) as bridging ligands (Fig. 1). Oxo ligands stabilize high oxidation states of a metal. They are also found in several metalloproteins, for example in molybdenum cofactors and in many iron-containing enzymes. One of the earliest synthetic compounds to incorporate an oxo ligand is potassium ferrate (K2FeO4), which was likely prepared by Georg E. Stahl in 1702.

Ferredoxin-thioredoxin reductase EC 1.8.7.2, systematic name ferredoxin:thioredoxin disulfide oxidoreductase, is a [4Fe-4S] protein that plays an important role in the ferredoxin/thioredoxin regulatory chain. It catalyzes the following reaction:
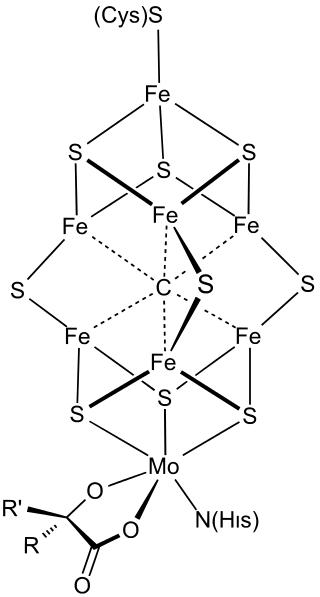
FeMoco (FeMo cofactor) is the primary cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Studying FeMoco's role in the reaction mechanism for nitrogen fixation is a potential use case for quantum computers. Even limited quantum computers could enable better simulations of the reaction mechanism.

In enzymology, an aldehyde ferredoxin oxidoreductase (EC 1.2.7.5) is an enzyme that catalyzes the chemical reaction
Evolution of metal ions in biological systems refers to the incorporation of metallic ions into living organisms and how it has changed over time. Metal ions have been associated with biological systems for billions of years, but only in the last century have scientists began to truly appreciate the scale of their influence. Major and minor metal ions have become aligned with living organisms through the interplay of biogeochemical weathering and metabolic pathways involving the products of that weathering. The associated complexes have evolved over time.
In enzymology, a formylmethanofuran dehydrogenase (EC 1.2.99.5) is an enzyme that catalyzes the chemical reaction:
![Overall structure of the [4FE-4S] cluster and the tungstoenzyme encompassed in the protein; the protein backbone is dark blue at the N-terminal end and green at the C-terminal end. Acetylene Hydratase from Pelobacter acetylenicus.png](http://upload.wikimedia.org/wikipedia/commons/thumb/6/63/Acetylene_Hydratase_from_Pelobacter_acetylenicus.png/250px-Acetylene_Hydratase_from_Pelobacter_acetylenicus.png)










