Related Research Articles
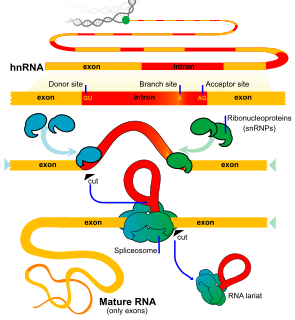
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA genes and noncoding DNA. Haploid human genomes, which are contained in germ cells consist of 3,054,815,472 DNA base pairs, while female diploid genomes have twice the DNA content.
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functions of non-coding DNA include the transcriptional and translational regulation of protein-coding sequences, scaffold attachment regions, origins of DNA replication, centromeres and telomeres. Its RNA counterpart is non-coding RNA.

Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by DNA duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for transcription or translation, or whose coding sequences are obviously defective due to frameshifts or premature stop codons.
Bacillus safensis is a Gram-positive, spore-forming, and rod bacterium, originally isolated from a spacecraft in Florida and California. B. safensis could have possibly been transported to the planet Mars on spacecraft Opportunity and Spirit in 2004. There are several known strains of this bacterium, all of which belong to the Bacillota phylum of Bacteria. This bacterium also belongs to the large, pervasive genus Bacillus. B. safensis is an aerobic chemoheterotroph and is highly resistant to salt and UV radiation. B. safensis affects plant growth, since it is a powerful plant hormone producer, and it also acts as a plant growth-promoting rhizobacteria, enhancing plant growth after root colonization. Strain B. safensis JPL-MERTA-8-2 is the only bacterial strain shown to grow noticeably faster in micro-gravity environments than on the Earth surface.
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.

In biology, a gene is a basic unit of heredity and a sequence of nucleotides in DNA that encodes the synthesis of a gene product, either RNA or protein.

60S ribosomal protein L22 is a protein that in humans is encoded by the RPL22 gene on Chromosome 1.

39S ribosomal protein L30, mitochondrial is a protein that in humans is encoded by the MRPL30 gene.

Tiling arrays are a subtype of microarray chips. Like traditional microarrays, they function by hybridizing labeled DNA or RNA target molecules to probes fixed onto a solid surface.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

28S ribosomal protein S21, mitochondrial is a protein that in humans is encoded by the MRPS21 gene.

HOTAIR is a human gene located between HOXC11 and HOXC12 on chromosome 12. It is the first example of an RNA expressed on one chromosome that has been found to influence transcription of HOXD cluster posterior genes located on chromosome 2. The sequence and function of HOTAIR is different in human and mouse. Sequence analysis of HOTAIR revealed that it exists in mammals, has poorly conserved sequences and considerably conserved structures, and has evolved faster than nearby HoxC genes. A subsequent study identified HOTAIR has 32 nucleotide long conserved noncoding element (CNE) that has a paralogous copy in HOXD cluster region, suggesting that the HOTAIR conserved sequences predates whole genome duplication events at the root of vertebrate. While the conserved sequence paralogous with HOXD cluster is 32 nucleotide long, the HOTAIR sequence conserved from human to fish is about 200 nucleotide long and is marked by active enhancer features.
Propionibacterium freudenreichii is a gram-positive, non-motile bacterium that plays an important role in the creation of Emmental cheese, and to some extent, Jarlsberg cheese, Leerdammer and Maasdam cheese. Its concentration in Swiss-type cheeses is higher than in any other cheese. Propionibacteria are commonly found in milk and dairy products, though they have also been extracted from soil. P. freudenreichii has a circular chromosome about 2.5 Mb long. When Emmental cheese is being produced, P. freudenreichii ferments lactate to form acetate, propionate, and carbon dioxide (3 C3H6O3 → 2 C2H5CO2 + C2H3O2 + CO2).
GENCODE is a scientific project in genome research and part of the ENCODE scale-up project.
A conserved non-coding sequence (CNS) is a DNA sequence of noncoding DNA that is evolutionarily conserved. These sequences are of interest for their potential to regulate gene production.
Rhodoferax is a genus of Betaproteobacteria belonging to the purple nonsulfur bacteriarophic. Originally, Rhodoferax species were included in the genus Rhodocyclus as the Rhodocyclus gelatinous-like group. The genus Rhodoferax was first proposed in 1991 to accommodate the taxonomic and phylogenetic discrepancies arising from its inclusion in the genus Rhodocyclus. Rhodoferax currently comprises four described species: R. fermentans, R. antarcticus, R. ferrireducens, and R. saidenbachensis. R. ferrireducens, lacks the typical phototrophic character common to two other Rhodoferax species. This difference has led researchers to propose the creation of a new genus, Albidoferax, to accommodate this divergent species. The genus name was later corrected to Albidiferax. Based on geno- and phenotypical characteristics, A. ferrireducens was reclassified in the genus Rhodoferax in 2014. R. saidenbachensis, a second non-phototrophic species of the genus Rhodoferax was described by Kaden et al. in 2014.
Lactiplantibacillus fabifermentans is a member of the genus Lactiplantibacillus and a type of lactic acid bacteria (LAB), a group of Gram-positive bacteria that produce lactic acid as their major fermented end product and that are often involved in food fermentation. L. fabifermentans was proposed in 2009 as a new species, after the type strain LMG 24284T has been isolated from Ghanaian cocoa fermentation. Analysis of the 16S rRNA gene sequence demonstrated that this species is a member of the Lactobacillus plantarum species group but further analysis demonstrated that it is possible to differentiate it from the nearest neighbors by means of DNA-DNA hybridization experiments, pheS sequence analysis, whole-cell protein electrophoresis, fluorescent amplified fragment length polymorphism analysis and biochemical characterization.
Halanaerobium hydrogeniformans is an alkaliphilic bacterium that is capable of biohydrogen production at and 33 °C (91 °F); it is commonly found in haloalkaline lakes.
Halostagnicola larsenii is a non-motile, aerobic, gram-negative, rod shaped archaeon. It is a halophilic, neutrophilic, chemo-organotroph and was isolated from samples taken from a saline lake in China. The etymology of the name comes from hals, halos Greek for salt, stagnum Latin for a piece of standing water, -cola Latin for inhabitant or dweller, and Larsenii named after the Norwegian microbiologist, Helge Larsen, who was a pioneer in research regarding halophiles.
References
- ↑ Parte, A.C. "Pelobacter". LPSN .
- 1 2 Schink, Bernhard (1985-08-01). "Fermentation of acetylene by an obligate anaerobe, Pelobacter acetylenicus sp. nov". Archives of Microbiology. 142 (3): 295–301. doi:10.1007/bf00693407. ISSN 0302-8933. S2CID 9331029.
- ↑ Sutton, John M.; Baesman, Shaun M.; Fierst, Janna L.; Poret-Peterson, Amisha T.; Oremland, Ronald S.; Dunlap, Darren S.; Akob, Denise M. (2017-02-09). "Complete Genome Sequences of Two Acetylene-Fermenting Pelobacter acetylenicus Strains". Genome Announcements. 5 (6). doi:10.1128/genomeA.01572-16. PMC 5331499 . PMID 28183759.