
Dijkstra's algorithm is an algorithm for finding the shortest paths between nodes in a weighted graph, which may represent, for example, road networks. It was conceived by computer scientist Edsger W. Dijkstra in 1956 and published three years later.

R-trees are tree data structures used for spatial access methods, i.e., for indexing multi-dimensional information such as geographical coordinates, rectangles or polygons. The R-tree was proposed by Antonin Guttman in 1984 and has found significant use in both theoretical and applied contexts. A common real-world usage for an R-tree might be to store spatial objects such as restaurant locations or the polygons that typical maps are made of: streets, buildings, outlines of lakes, coastlines, etc. and then find answers quickly to queries such as "Find all museums within 2 km of my current location", "retrieve all road segments within 2 km of my location" or "find the nearest gas station". The R-tree can also accelerate nearest neighbor search for various distance metrics, including great-circle distance.
A B+ tree is an m-ary tree with a variable but often large number of children per node. A B+ tree consists of a root, internal nodes and leaves. The root may be either a leaf or a node with two or more children.
In computer science, an interval tree is a tree data structure to hold intervals. Specifically, it allows one to efficiently find all intervals that overlap with any given interval or point. It is often used for windowing queries, for instance, to find all roads on a computerized map inside a rectangular viewport, or to find all visible elements inside a three-dimensional scene. A similar data structure is the segment tree.

In computer science, a k-d tree is a space-partitioning data structure for organizing points in a k-dimensional space. K-dimensional is that which concerns exactly k orthogonal axes or a space of any number of dimensions. k-d trees are a useful data structure for several applications, such as:
In statistics, the k-nearest neighbors algorithm (k-NN) is a non-parametric supervised learning method first developed by Evelyn Fix and Joseph Hodges in 1951, and later expanded by Thomas Cover. It is used for classification and regression. In both cases, the input consists of the k closest training examples in a data set. The output depends on whether k-NN is used for classification or regression:
A vantage-point tree is a metric tree that segregates data in a metric space by choosing a position in the space and partitioning the data points into two parts: those points that are nearer to the vantage point than a threshold, and those points that are not. By recursively applying this procedure to partition the data into smaller and smaller sets, a tree data structure is created where neighbors in the tree are likely to be neighbors in the space.
Nearest neighbor search (NNS), as a form of proximity search, is the optimization problem of finding the point in a given set that is closest to a given point. Closeness is typically expressed in terms of a dissimilarity function: the less similar the objects, the larger the function values.
In computer science, fractional cascading is a technique to speed up a sequence of binary searches for the same value in a sequence of related data structures. The first binary search in the sequence takes a logarithmic amount of time, as is standard for binary searches, but successive searches in the sequence are faster. The original version of fractional cascading, introduced in two papers by Chazelle and Guibas in 1986, combined the idea of cascading, originating in range searching data structures of Lueker (1978) and Willard (1978), with the idea of fractional sampling, which originated in Chazelle (1983). Later authors introduced more complex forms of fractional cascading that allow the data structure to be maintained as the data changes by a sequence of discrete insertion and deletion events.
In computer science, locality-sensitive hashing (LSH) is a fuzzy hashing technique that hashes similar input items into the same "buckets" with high probability. Since similar items end up in the same buckets, this technique can be used for data clustering and nearest neighbor search. It differs from conventional hashing techniques in that hash collisions are maximized, not minimized. Alternatively, the technique can be seen as a way to reduce the dimensionality of high-dimensional data; high-dimensional input items can be reduced to low-dimensional versions while preserving relative distances between items.

Density-based spatial clustering of applications with noise (DBSCAN) is a data clustering algorithm proposed by Martin Ester, Hans-Peter Kriegel, Jörg Sander and Xiaowei Xu in 1996. It is a density-based clustering non-parametric algorithm: given a set of points in some space, it groups together points that are closely packed together, marking as outliers points that lie alone in low-density regions . DBSCAN is one of the most common, and most commonly cited, clustering algorithms.
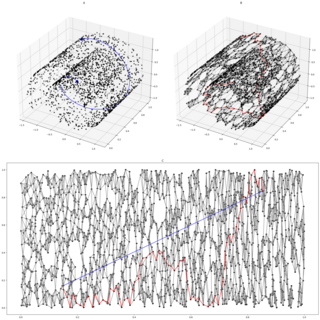
Isomap is a nonlinear dimensionality reduction method. It is one of several widely used low-dimensional embedding methods. Isomap is used for computing a quasi-isometric, low-dimensional embedding of a set of high-dimensional data points. The algorithm provides a simple method for estimating the intrinsic geometry of a data manifold based on a rough estimate of each data point’s neighbors on the manifold. Isomap is highly efficient and generally applicable to a broad range of data sources and dimensionalities.
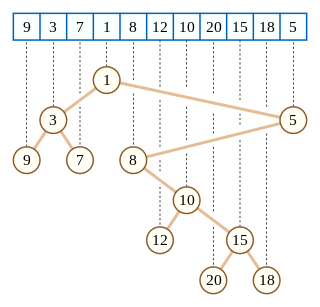
In computer science, a Cartesian tree is a binary tree derived from a sequence of distinct numbers. To construct the Cartesian tree, set its root to be the minimum number in the sequence, and recursively construct its left and right subtrees from the subsequences before and after this number. It is uniquely defined as a min-heap whose symmetric (in-order) traversal returns the original sequence.
In pattern recognition, the iDistance is an indexing and query processing technique for k-nearest neighbor queries on point data in multi-dimensional metric spaces. The kNN query is one of the hardest problems on multi-dimensional data, especially when the dimensionality of the data is high. The iDistance is designed to process kNN queries in high-dimensional spaces efficiently and it is especially good for skewed data distributions, which usually occur in real-life data sets. The iDistance can be augmented with machine learning models to learn the data distributions for searching and storing the multi-dimensional data.
In computer science, the Bx tree is a query that is used to update efficient B+ tree-based index structures for moving objects.
In computer science, M-trees are tree data structures that are similar to R-trees and B-trees. It is constructed using a metric and relies on the triangle inequality for efficient range and k-nearest neighbor (k-NN) queries. While M-trees can perform well in many conditions, the tree can also have large overlap and there is no clear strategy on how to best avoid overlap. In addition, it can only be used for distance functions that satisfy the triangle inequality, while many advanced dissimilarity functions used in information retrieval do not satisfy this.
In the theory of cluster analysis, the nearest-neighbor chain algorithm is an algorithm that can speed up several methods for agglomerative hierarchical clustering. These are methods that take a collection of points as input, and create a hierarchy of clusters of points by repeatedly merging pairs of smaller clusters to form larger clusters. The clustering methods that the nearest-neighbor chain algorithm can be used for include Ward's method, complete-linkage clustering, and single-linkage clustering; these all work by repeatedly merging the closest two clusters but use different definitions of the distance between clusters. The cluster distances for which the nearest-neighbor chain algorithm works are called reducible and are characterized by a simple inequality among certain cluster distances.
In computational geometry, a well-separated pair decomposition (WSPD) of a set of points , is a sequence of pairs of sets , such that each pair is well-separated, and for each two distinct points , there exists precisely one pair which separates the two.
The PH-tree is a tree data structure used for spatial indexing of multi-dimensional data (keys) such as geographical coordinates, points, feature vectors, rectangles or bounding boxes. The PH-tree is space partitioning index with a structure similar to that of a quadtree or octree. However, unlike quadtrees, it uses a splitting policy based on tries and similar to Crit bit trees that is based on the bit-representation of the keys. The bit-based splitting policy, when combined with the use of different internal representations for nodes, provides scalability with high-dimensional data. The bit-representation splitting policy also imposes a maximum depth, thus avoiding degenerated trees and the need for rebalancing.
The compressed cover tree is a type of data structure in computer science that is specifically designed to facilitate the speed-up of a k-nearest neighbors algorithm in finite metric spaces. Compressed cover tree is a simplified version of explicit representation of cover tree that was motivated by past issues in proofs of time complexity results of cover tree. The compressed cover tree was specifically designed to achieve claimed time complexities of cover tree in a mathematically rigorous way.










