Related Research Articles
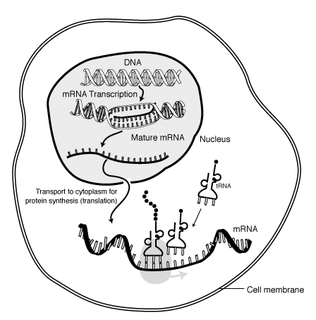
Messenger RNA (mRNA) is a single-stranded RNA molecule that corresponds to the genetic sequence of a gene and is read by the ribosome in the process of producing a protein. mRNA is created during the process of transcription, where the enzyme RNA polymerase converts genes into primary transcript mRNA. This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system.
In molecular biology, the five-prime cap is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped.
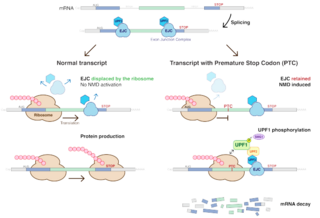
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins.
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation. Several initiation factors form a complex with the small 40S ribosomal subunit and Met-tRNAiMet called the 43S preinitiation complex. Additional factors of the eIF4F complex recruit the 43S PIC to the five-prime cap structure of the mRNA, from which the 43S particle scans 5'-->3' along the mRNA to reach an AUG start codon. Recognition of the start codon by the Met-tRNAiMet promotes gated phosphate and eIF1 release to form the 48S preinitiation complex, followed by large 60S ribosomal subunit recruitment to form the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors, reflecting the greater biological complexity of eukaryotic translation. There are at least twelve eukaryotic initiation factors, composed of many more polypeptides, and these are described below.

RNA-binding protein 8A is a protein that in humans is encoded by the RBM8A gene.
Regulator of nonsense transcripts 1 is a protein that in humans is encoded by the UPF1 gene.

Regulator of nonsense transcripts 2 is a protein that in humans is encoded by the UPF2 gene.

Regulator of nonsense transcripts 3B is a protein that in humans is encoded by the UPF3B gene.

Eukaryotic translation initiation factor 4 gamma 3 is a protein that in humans is encoded by the EIF4G3 gene. The gene encodes a protein that functions in translation by aiding the assembly of the ribosome onto the messenger RNA template. Confusingly, this protein is usually referred to as eIF4GII, as although EIF4G3 is the third gene that is similar to eukaryotic translation initiation factor 4 gamma, the second isoform EIF4G2 is not an active translation initiation factor.

mRNA-decapping enzyme 2 is a protein that in humans is encoded by the DCP2 gene.

Regulator of nonsense transcripts 3A is a protein that in humans is encoded by the UPF3A gene.

Scavenger mRNA-decapping enzyme DcpS is a protein that in humans is encoded by the DCPS gene.

mRNA-decapping enzyme 1A is a protein that in humans is encoded by the DCP1A gene.

Enhancer of mRNA-decapping protein 3 is a protein that in humans is encoded by the EDC3 gene.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human eIF4G 1 has been the focus of extensive studies.
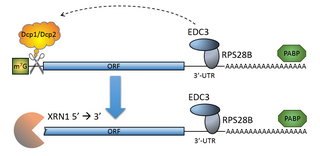
The process of messenger RNA decapping consists of hydrolysis of the 5' cap structure on the RNA exposing a 5' monophosphate. In eukaryotes, this 5' monophosphate is a substrate for the 5' exonuclease Xrn1 and the mRNA is quickly destroyed. There are many situations which may lead to the removal of the cap, some of which are discussed below.
The mRNA decapping complex is a protein complex in eukaryotic cells responsible for removal of the 5' cap. The active enzyme of the decapping complex is the bilobed Nudix family enzyme Dcp2, which hydrolyzes 5' cap and releases 7mGDP and a 5'-monophosphorylated mRNA. This decapped mRNA is inhibited for translation and will be degraded by exonucleases. The core decapping complex is conserved in eukaryotes. Dcp2 is activated by Decapping Protein 1 (Dcp1) and in higher eukaryotes joined by the scaffold protein VCS. Together with many other accessory proteins, the decapping complex assembles in P-bodies in the cytoplasm.
mRNA surveillance mechanisms are pathways utilized by organisms to ensure fidelity and quality of messenger RNA (mRNA) molecules. There are a number of surveillance mechanisms present within cells. These mechanisms function at various steps of the mRNA biogenesis pathway to detect and degrade transcripts that have not properly been processed.
An exon junction complex (EJC) is a protein complex which forms on a pre-messenger RNA strand at the junction of two exons which have been joined together during RNA splicing. The EJC has major influences on translation, surveillance and localization of the spliced mRNA. It is first deposited onto mRNA during splicing and is then transported into the cytoplasm. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that exon junction complexes provide a position-specific memory of the splicing event. The EJC consists of a stable heterotetramer core, which serves as a binding platform for other factors necessary for the mRNA pathway. The core of the EJC contains the protein eukaryotic initiation factor 4A-III bound to an adenosine triphosphate (ATP) analog, as well as the additional proteins Magoh and Y14.The binding of these proteins to nuclear speckled domains has been measured recently and it may be regulated by PI3K/AKT/mTOR signaling pathways. In order for the binding of the complex to the mRNA to occur, the eIF4AIII factor is inhibited, stopping the hydrolysis of ATP. This recognizes EJC as an ATP dependent complex. EJC also interacts with a large number of additional proteins; most notably SR proteins. These interactions are suggested to be important for mRNA compaction. The role of EJC in mRNA export is controversial.
M7GpppN-mRNA hydrolase (EC 3.6.1.62, DCP2, NUDT16, D10 protein, D9 protein, D10 decapping enzyme, decapping enzyme) is an enzyme with systematic name m7GpppN-mRNA m7GDP phosphohydrolase. This enzyme catalyses the following chemical reaction
References
- ↑ Isken, O.; Maquat, L.E. (2007), "Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function", Genes & Development, 21 (15): 1833–56, doi: 10.1101/gad.1566807 , PMID 17671086
- ↑ Maquat, Lynne E.; Tarn, Woan-Yuh; Isken, Olaf (August 2010). "The Pioneer Round of Translation: Features and Functions". Cell. 142 (3): 368–374. doi:10.1016/j.cell.2010.07.022. PMC 2950652 . PMID 20691898.
- ↑ Gross, J.D.; Moerke, N.J.; Von Der Haar, T.; Lugovskoy, A.A.; Sachs, A.B.; McCarthy, J.E.G.; Wagner, G. (2003), "Ribosome Loading onto the mRNA Cap is Driven by Conformational Coupling between eIF4G and eIF4E", Cell, 115 (6): 739–750, doi:10.1016/S0092-8674(03)00975-9, PMID 14675538
- ↑ Parker, R.; Sheth, U. (2007), "P Bodies and the Control of mRNA Translation and Degradation", Molecular Cell, 25 (5): 635–646, doi:10.1016/j.molcel.2007.02.011, PMID 17349952