
The thymus is a specialized primary lymphoid organ of the immune system. Within the thymus, thymus cell lymphocytes or T cells mature. T cells are critical to the adaptive immune system, where the body adapts to specific foreign invaders. The thymus is located in the upper front part of the chest, in the anterior superior mediastinum, behind the sternum, and in front of the heart. It is made up of two lobes, each consisting of a central medulla and an outer cortex, surrounded by a capsule.

T cells are one of the important types of white blood cells of the immune system and play a central role in the adaptive immune response. T cells can be distinguished from other lymphocytes by the presence of a T-cell receptor (TCR) on their cell surface.

A cytotoxic T cell (also known as TC, cytotoxic T lymphocyte, CTL, T-killer cell, cytolytic T cell, CD8+ T-cell or killer T cell) is a T lymphocyte (a type of white blood cell) that kills cancer cells, cells that are infected by intracellular pathogens (such as viruses or bacteria), or cells that are damaged in other ways.
The regulatory T cells (Tregs or Treg cells), formerly known as suppressor T cells, are a subpopulation of T cells that modulate the immune system, maintain tolerance to self-antigens, and prevent autoimmune disease. Treg cells are immunosuppressive and generally suppress or downregulate induction and proliferation of effector T cells. Treg cells express the biomarkers CD4, FOXP3, and CD25 and are thought to be derived from the same lineage as naïve CD4+ cells. Because effector T cells also express CD4 and CD25, Treg cells are very difficult to effectively discern from effector CD4+, making them difficult to study. Research has found that the cytokine transforming growth factor beta (TGF-β) is essential for Treg cells to differentiate from naïve CD4+ cells and is important in maintaining Treg cell homeostasis.
Cross-presentation is the ability of certain professional antigen-presenting cells (mostly dendritic cells) to take up, process and present extracellular antigens with MHC class I molecules to CD8 T cells (cytotoxic T cells). Cross-priming, the result of this process, describes the stimulation of naive cytotoxic CD8+ T cells into activated cytotoxic CD8+ T cells. This process is necessary for immunity against most tumors and against viruses that infect dendritic cells and sabotage their presentation of virus antigens. Cross presentation is also required for the induction of cytotoxic immunity by vaccination with protein antigens, for example, tumour vaccination.
In immunology, central tolerance is the process of eliminating any developing T or B lymphocytes that are autoreactive, i.e. reactive to the body itself. Through elimination of autoreactive lymphocytes, tolerance ensures that the immune system does not attack self peptides. Lymphocyte maturation occurs in primary lymphoid organs such as the bone marrow and the thymus. In mammals, B cells mature in the bone marrow and T cells mature in the thymus.
A thymocyte is an immune cell present in the thymus, before it undergoes transformation into a T cell. Thymocytes are produced as stem cells in the bone marrow and reach the thymus via the blood.

Antigen presentation is a vital immune process that is essential for T cell immune response triggering. Because T cells recognize only fragmented antigens displayed on cell surfaces, antigen processing must occur before the antigen fragment can be recognized by a T-cell receptor. Specifically, the fragment, bound to the major histocompatibility complex (MHC), is transported to the surface of the cell, a process known as presentation. If there has been an infection with viruses or bacteria, the cell will present an endogenous or exogenous peptide fragment derived from the antigen by MHC molecules. There are two types of MHC molecules which differ in the behaviour of the antigens: MHC class I molecules (MHC-I) bind peptides from the cell cytosol, while peptides generated in the endocytic vesicles after internalisation are bound to MHC class II (MHC-II). Cellular membranes separate these two cellular environments - intracellular and extracellular. Each T cell can only recognize tens to hundreds of copies of a unique sequence of a single peptide among thousands of other peptides presented on the same cell, because an MHC molecule in one cell can bind to quite a large range of peptides. Predicting which antigens will be presented to the immune system by a certain MHC/HLA type is difficult, but the technology involved is improving.
MHC-restricted antigen recognition, or MHC restriction, refers to the fact that a T cell can interact with a self-major histocompatibility complex molecule and a foreign peptide bound to it, but will only respond to the antigen when it is bound to a particular MHC molecule.
Self-protein refers to all proteins endogenously produced by DNA-level transcription and translation within an organism of interest. This does not include proteins synthesized due to viral infection, but may include those synthesized by commensal bacteria within the intestines. Proteins that are not created within the body of the organism of interest, but nevertheless enter through the bloodstream, a breach in the skin, or a mucous membrane, may be designated as “non-self” and subsequently targeted and attacked by the immune system. Tolerance to self-protein is crucial for overall wellbeing; when the body erroneously identifies self-proteins as “non-self”, the subsequent immune response against endogenous proteins may lead to the development of an autoimmune disease.

Minor histocompatibility antigen are peptides presented on the cellular surface of donated organs that are known to give an immunological response in some organ transplants. They cause problems of rejection less frequently than those of the major histocompatibility complex (MHC). Minor histocompatibility antigens (MiHAs) are diverse, short segments of proteins and are referred to as peptides. These peptides are normally around 9-12 amino acids in length and are bound to both the major histocompatibility complex (MHC) class I and class II proteins. Peptide sequences can differ among individuals and these differences arise from SNPs in the coding region of genes, gene deletions, frameshift mutations, or insertions. About a third of the characterized MiHAs come from the Y chromosome. Prior to becoming a short peptide sequence, the proteins expressed by these polymorphic or diverse genes need to be digested in the proteasome into shorter peptides. These endogenous or self peptides are then transported into the endoplasmic reticulum with a peptide transporter pump called TAP where they encounter and bind to the MHC class I molecule. This contrasts with MHC class II molecules's antigens which are peptides derived from phagocytosis/endocytosis and molecular degradation of non-self entities' proteins, usually by antigen-presenting cells. MiHA antigens are either ubiquitously expressed in most tissue like skin and intestines or restrictively expressed in the immune cells.
In immunology, peripheral tolerance is the second branch of immunological tolerance, after central tolerance. It takes place in the immune periphery. Its main purpose is to ensure that self-reactive T and B cells which escaped central tolerance do not cause autoimmune disease. Peripheral tolerance prevents immune response to harmless food antigens and allergens, too.
In immunology, clonal deletion is the removal through apoptosis of B cells and T cells that have expressed receptors for self before developing into fully immunocompetent lymphocytes. This prevents recognition and destruction of self host cells, making it a type of negative selection or central tolerance. Central tolerance prevents B and T lymphocytes from reacting to self. Thus, clonal deletion can help protect individuals against autoimmunity. Clonal deletion is thought to be the most common type of negative selection. It is one method of immune tolerance.
Thymic nurse cells (TNCs) are large epithelial cells found in the cortex of the thymus and also in cortico-medullary junction. They have their own nucleus and are known to internalize thymocytes through extensions of plasma membrane. The cell surfaces of TNCs and their cytoplasmic vacuoles express MHC Class I and MHC Class II antigens. The interaction of these antigens with the developing thymocytes determines whether the thymocytes undergo positive or negative selection.
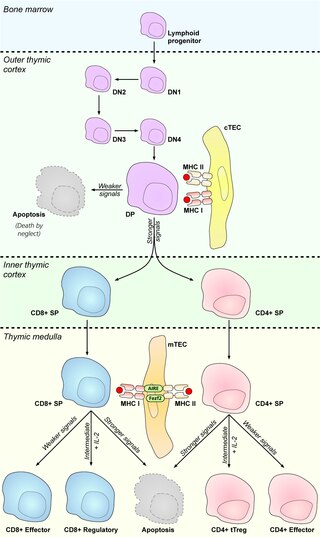
Medullary thymic epithelial cells (mTECs) represent a unique stromal cell population of the thymus which plays an essential role in the establishment of central tolerance. Therefore, mTECs rank among cells relevant for the development of functional mammal immune system.
Antigen transfer in the thymus is the transmission of self-antigens between thymic antigen-presenting cells which contributes to the establishment of T cell central tolerance.
Thymic epithelial cells (TECs) are specialized cells with high degree of anatomic, phenotypic and functional heterogeneity that are located in the outer layer (epithelium) of the thymic stroma. The thymus, as a primary lymphoid organ, mediates T cell development and maturation. The thymic microenvironment is established by TEC network filled with thymocytes in different developing stages. TECs and thymocytes are the most important components in the thymus, that are necessary for production of functionally competent T lymphocytes and self tolerance. Dysfunction of TECs causes several immunodeficiencies and autoimmune diseases.
Promiscuous gene expression (PGE), formerly referred to as ectopic expression, is a process specific to the thymus that plays a pivotal role in the establishment of central tolerance. This phenomenon enables generation of self-antigens, so called tissue-restricted antigens (TRAs), which are in the body expressed only by one or few specific tissues. These antigens are represented for example by insulin from the pancreas or defensins from the gastrointestinal tract. Antigen-presenting cells (APCs) of the thymus, namely medullary thymic epithelial cells (mTECs), dendritic cells (DCs) and B cells are capable to present peptides derived from TRAs to developing T cells and hereby test, whether their T cell receptors (TCRs) engage self entities and therefore their occurrence in the body can potentially lead to the development of autoimmune disease. In that case, thymic APCs either induce apoptosis in these autoreactive T cells or they deviate them to become T regulatory cells, which suppress self-reactive T cells in the body that escaped negative selection in the thymus. Thus, PGE is crucial for tissues protection against autoimmunity.
Thymoproteasome is a special kind of proteasome, which is present in vertebrates. In the body it is located in thymus, exclusively in cortical thymic epithelial cells (cTECs). But in thymus we can also find another type of specific proteasome, immunoproteasome, which is present in thymocytes, dendritic cells and medular thymic epithelial cells. It was first described in 2007 during a search for non-intronic sequence proximal to PSMB5 locus in mouse genome. The PSMB5 locus encodes the standard β5 proteasome subunit, while this sequence encodes a variant subunit β5t (PSMB11) specific to thymoproteasome. The importance of this protein complex is its involvement in positive selection of T cells.
Thymus stromal cells are subsets of specialized cells located in different areas of the thymus. They include all non-T-lineage cells, such as thymic epithelial cells (TECs), endothelial cells, mesenchymal cells, dendritic cells, and B lymphocytes, and provide signals essential for thymocyte development and the homeostasis of the thymic stroma.





