
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the positions of the atoms in the crystal can be determined, as well as their chemical bonds, crystallographic disorder, and various other information.
In physics, the phase problem is the problem of loss of information concerning the phase that can occur when making a physical measurement. The name comes from the field of X-ray crystallography, where the phase problem has to be solved for the determination of a structure from diffraction data. The phase problem is also met in the fields of imaging and signal processing. Various approaches of phase retrieval have been developed over the years.
Electron crystallography is a method to determine the arrangement of atoms in solids using a transmission electron microscope (TEM). It can involve the use of high-resolution transmission electron microscopy images, electron diffraction patterns including convergent-beam electron diffraction or combinations of these. It has been successful in determining some bulk structures, and also surface structures. Two related methods are low-energy electron diffraction which has solved the structure of many surfaces, and reflection high-energy electron diffraction which is used to monitor surfaces often during growth.
Hugo M. Rietveld was a Dutch crystallographer who is famous for his publication on the full profile refinement method in powder diffraction, which became later known as the Rietveld refinement method. The method is used for the characterisation of crystalline materials from X-ray powder diffraction data. The Rietveld refinement uses a least squares approach to refine a theoretical line profile until it matches the measured profile. The introduction of this technique which used the full profile instead of individual reflections was a significant step forward in the diffraction analysis of powder samples.
In X-ray crystallography, a difference density map or Fo–Fc map shows the spatial distribution of the difference between the measured electron density of the crystal and the electron density explained by the current model.
Multi-wavelength anomalous diffraction is a technique used in X-ray crystallography that facilitates the determination of the three-dimensional structure of biological macromolecules via solution of the phase problem.
Acta Crystallographica is a series of peer-reviewed scientific journals, with articles centred on crystallography, published by the International Union of Crystallography (IUCr). Originally established in 1948 as a single journal called Acta Crystallographica, there are now six independent Acta Crystallographica titles:
Isomorphous replacement (IR) is historically the most common approach to solving the phase problem in X-ray crystallography studies of proteins. For protein crystals this method is conducted by soaking the crystal of a sample to be analyzed with a heavy atom solution or co-crystallization with the heavy atom. The addition of the heavy atom (or ion) to the structure should not affect the crystal formation or unit cell dimensions in comparison to its native form, hence, they should be isomorphic.
A crystallographic database is a database specifically designed to store information about the structure of molecules and crystals. Crystals are solids having, in all three dimensions of space, a regularly repeating arrangement of atoms, ions, or molecules. They are characterized by symmetry, morphology, and directionally dependent physical properties. A crystal structure describes the arrangement of atoms, ions, or molecules in a crystal.
Eleanor Joy Dodson FRS is an Australian-born biologist who specialises in the computational modelling of protein crystallography. She holds a chair in the Department of Chemistry at the University of York. She is the widow of the scientist Guy Dodson.
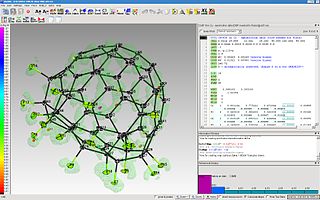
The program ShelXle is a graphical user interface for the structure refinement program SHELXL. ShelXle combines an editor with syntax highlighting for the SHELXL-associated .ins (input) and .res (output) files with an interactive graphical display for visualization of a three-dimensional structure including the electron density (Fo) and difference density (Fo-Fc) maps.

Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule, come from structural biology experiments such as x-ray crystallography or nuclear magnetic resonance (NMR). The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.
George Michael Sheldrick, FRS is a British chemist who specialises in molecular structure determination. He is one of the most cited workers in the field, having over 280,000 citations as of 2020 and an h-index of 113. He was a professor at the University of Göttingen from 1978 until his retirement in 2011.
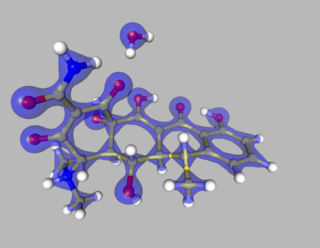
The Multipole Density Formalism is an X-ray crystallography method of electron density modelling proposed by Niels K. Hansen and Philip Coppens in 1978. Unlike the commonly used Independent Atom Model, the Hansen-Coppens Formalism presents an aspherical approach, allowing one to model the electron distribution around a nucleus separately in different directions and therefore describe numerous chemical features of a molecule inside the unit cell of an examined crystal in detail.
Microcrystal electron diffraction, or MicroED, is a CryoEM method that was developed by the Gonen laboratory in late 2013 at the Janelia Research Campus of the Howard Hughes Medical Institute. MicroED is a form of electron crystallography where thin 3D crystals are used for structure determination by electron diffraction. Prior to this demonstration, macromolecular (protein) electron crystallography was only used on 2D crystals, for example.
Aafje Looijenga-Vos was a Dutch crystallographer. She was a professor for general chemistry and later for structural chemistry at the University of Groningen.

Durward William John Cruickshank, often known as D. W. J. Cruickshank, was a British crystallographer whose work transformed the precision of determining molecular structures from X-ray crystal structure analysis. He developed the theoretical framework for anisotropic displacement parameters, also known as the thermal ellipsoid, for crystal structure determination in a series of papers published in 1956 in Acta Crystallographica.
This is a timeline of crystallography.
Hans-Beat Bürgi is a Swiss chemist and crystallographer. He was a professor for crystallography at the University of Bern from 1979 to 2007.
Urea can crystallise with other compounds. These can be called urea adducts or if a solvent is involved, a urea solvate, and the process is called urea extraction crystallization. Urea can also be a neutral ligand if it is coordinated to a central metal atom. Urea can form hydrogen bonds to other oxygen and nitrogen atoms in the substance it crystallises with. This stiffens the solid and raises the melting point. T





