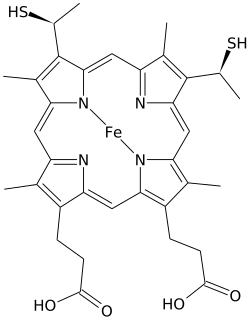
Cytochromes are redox-active proteins containing a heme, with a central iron (Fe) atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d.
In biochemistry, a disulfide refers to a functional group with the structure R−S−S−R′. The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. Persulfide usually refers to R−S−S−H compounds.

The cytochrome complex, or cyt c, is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion where it plays a critical role in cellular respiration. It transfers electrons between Complexes III and IV. Cytochrome c is highly water-soluble, unlike other cytochromes. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It also plays a major role in cell apoptosis. In humans, cytochrome c is encoded by the CYCS gene.

A hemeprotein, or heme protein, is a protein that contains a heme prosthetic group. They are a very large class of metalloproteins. The heme group confers functionality, which can include oxygen carrying, oxygen reduction, electron transfer, and other processes. Heme is bound to the protein either covalently or noncovalently or both.

Heme, or haem, is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.

Post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes translating mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.

Cytochrome c peroxidase, or CCP, is a water-soluble heme-containing enzyme of the peroxidase family that takes reducing equivalents from cytochrome c and reduces hydrogen peroxide to water:
A prosthetic group is the non-amino acid component that is part of the structure of the heteroproteins or conjugated proteins, being tightly linked to the apoprotein.

Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.

Cysteine dioxygenase (CDO) is a non-heme iron enzyme that catalyzes the conversion of L-cysteine to cysteine sulfinic acid. CDO plays an important role in cysteine catabolism, regulating intracellular levels of cysteine and responding changes in cysteine availability. As such, CDO is highly regulated and undergoes large changes in concentration and efficiency. It oxidizes cysteine to the corresponding sulfinic acid by activation of dioxygen, although the exact mechanism of the reaction is still unclear. In addition to being found in mammals, CDO also exists in some yeast and bacteria, although the exact function is still unknown. CDO has been implicated in various neurodegenerative diseases and cancers, which is likely related to cysteine toxicity.

Rieske proteins are iron–sulfur protein (ISP) components of cytochrome bc1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein
It is a unique [2Fe-2S] cluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV.

Thromboxane A synthase 1 , also known as TBXAS1, is a cytochrome P450 enzyme that, in humans, is encoded by the TBXAS1 gene.
Copper proteins are proteins that contain one or more copper ions as prosthetic groups. Copper proteins are found in all forms of air-breathing life. These proteins are usually associated with electron-transfer with or without the involvement of oxygen (O2). Some organisms even use copper proteins to carry oxygen instead of iron proteins. A prominent copper proteins in humans is in cytochrome c oxidase (cco). The enzyme cco mediates the controlled combustion that produces ATP.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
Iron-binding proteins are carrier proteins and metalloproteins that are important in iron metabolism and the immune response. Iron is required for life.
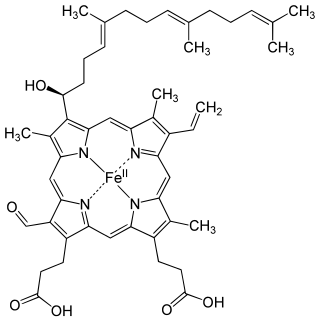
Heme A is a heme, a coordination complex consisting of a macrocyclic ligand called a porphyrin, chelating an iron atom. Heme A is a biomolecule and is produced naturally by many organisms. Heme A, often appears a dichroic green/red when in solution, is a structural relative of heme B, a component of hemoglobin, the red pigment in blood.

Cytochromes c cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds. These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage, a lysine or a methionine instead of the axial histidine or a CXnCH binding motif with n>2. The second axial site of the iron can be coordinated by amino acids of the protein, substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes. A prominent member of this family is mitochondrial cytochrome c.
Cytochrome c1, heme protein, mitochondrial (CYC1), also known as UQCR4, MC3DN6, Complex III subunit 4, Cytochrome b-c1 complex subunit 4, or Ubiquinol-cytochrome-c reductase complex cytochrome c1 subunit is a protein that in humans is encoded by the CYC1 gene. CYC1 is a respiratory subunit of Ubiquinol Cytochrome c Reductase, which is located in the inner mitochondrial membrane and is part of the electron transport chain. Mutations in this gene may cause mitochondrial complex III deficiency, nuclear, type 6.

Eosinophil peroxidase is an enzyme found within the eosinophil granulocytes, innate immune cells of humans and mammals. This oxidoreductase protein is encoded by the gene EPX, expressed within these myeloid cells. EPO shares many similarities with its orthologous peroxidases, myeloperoxidase (MPO), lactoperoxidase (LPO), and thyroid peroxidase (TPO). The protein is concentrated in secretory granules within eosinophils. Eosinophil peroxidase is a heme peroxidase, its activities including the oxidation of halide ions to bacteriocidal reactive oxygen species, the cationic disruption of bacterial cell walls, and the post-translational modification of protein amino acid residues.