Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in numerous biological pathways, including digestion of ingested proteins, protein catabolism, and cell signaling.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

Angiotensin-converting enzyme, or ACE, is a central component of the renin–angiotensin system (RAS), which controls blood pressure by regulating the volume of fluids in the body. It converts the hormone angiotensin I to the active vasoconstrictor angiotensin II. Therefore, ACE indirectly increases blood pressure by causing blood vessels to constrict. ACE inhibitors are widely used as pharmaceutical drugs for treatment of cardiovascular diseases.

Amylin, or islet amyloid polypeptide (IAPP), is a 37-residue peptide hormone. It is co-secreted with insulin from the pancreatic β-cells in the ratio of approximately 100:1 (insulin:amylin). Amylin plays a role in glycemic regulation by slowing gastric emptying and promoting satiety, thereby preventing post-prandial spikes in blood glucose levels.

Cathepsins are proteases found in all animals as well as other organisms. There are approximately a dozen members of this family, which are distinguished by their structure, catalytic mechanism, and which proteins they cleave. Most of the members become activated at the low pH found in lysosomes. Thus, the activity of this family lies almost entirely within those organelles. There are, however, exceptions such as cathepsin K, which works extracellularly after secretion by osteoclasts in bone resorption. Cathepsins have a vital role in mammalian cellular turnover.

Amyloid beta denotes peptides of 36–43 amino acids that are the main component of the amyloid plaques found in the brains of people with Alzheimer's disease. The peptides derive from the amyloid-beta precursor protein (APP), which is cleaved by beta secretase and gamma secretase to yield Aβ in a cholesterol-dependent process and substrate presentation. Aβ molecules can aggregate to form flexible soluble oligomers which may exist in several forms. It is now believed that certain misfolded oligomers can induce other Aβ molecules to also take the misfolded oligomeric form, leading to a chain reaction akin to a prion infection. The oligomers are toxic to nerve cells. The other protein implicated in Alzheimer's disease, tau protein, also forms such prion-like misfolded oligomers, and there is some evidence that misfolded Aβ can induce tau to misfold.

Amyloid-beta precursor protein (APP) is an integral membrane protein expressed in many tissues and concentrated in the synapses of neurons. It functions as a cell surface receptor and has been implicated as a regulator of synapse formation, neural plasticity, antimicrobial activity, and iron export. It is coded for by the gene APP and regulated by substrate presentation. APP is best known as the precursor molecule whose proteolysis generates amyloid beta (Aβ), a polypeptide containing 37 to 49 amino acid residues, whose amyloid fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients.
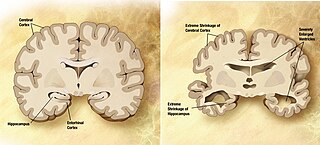
A neurodegenerative disease is caused by the progressive loss of structure or function of neurons, in the process known as neurodegeneration. Such neuronal damage may ultimately involve cell death. Neurodegenerative diseases include amyotrophic lateral sclerosis, multiple sclerosis, Parkinson's disease, Alzheimer's disease, Huntington's disease, multiple system atrophy, tauopathies, and prion diseases. Neurodegeneration can be found in the brain at many different levels of neuronal circuitry, ranging from molecular to systemic. Because there is no known way to reverse the progressive degeneration of neurons, these diseases are considered to be incurable; however research has shown that the two major contributing factors to neurodegeneration are oxidative stress and inflammation. Biomedical research has revealed many similarities between these diseases at the subcellular level, including atypical protein assemblies and induced cell death. These similarities suggest that therapeutic advances against one neurodegenerative disease might ameliorate other diseases as well.

Beta-secretase 1, also known as beta-site amyloid precursor protein cleaving enzyme 1, beta-site APP cleaving enzyme 1 (BACE1), membrane-associated aspartic protease 2, memapsin-2, aspartyl protease 2, and ASP2, is an enzyme that in humans is encoded by the BACE1 gene. Expression of BACE1 is observed mainly in neurons.
The biochemistry of Alzheimer's disease, the most common cause of dementia, is not yet very well understood. Alzheimer's disease (AD) has been identified as a proteopathy: a protein misfolding disease due to the accumulation of abnormally folded amyloid beta (Aβ) protein in the brain. Amyloid beta is a short peptide that is an abnormal proteolytic byproduct of the transmembrane protein amyloid-beta precursor protein (APP), whose function is unclear but thought to be involved in neuronal development. The presenilins are components of proteolytic complex involved in APP processing and degradation.
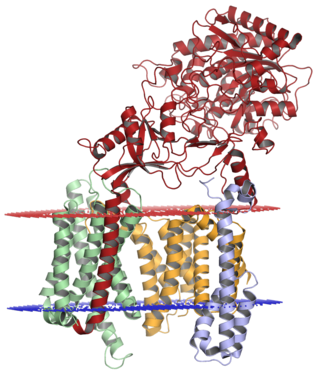
Gamma secretase is a multi-subunit protease complex, itself an integral membrane protein, that cleaves single-pass transmembrane proteins at residues within the transmembrane domain. Proteases of this type are known as intramembrane proteases. The most well-known substrate of gamma secretase is amyloid precursor protein, a large integral membrane protein that, when cleaved by both gamma and beta secretase, produces a short 37-43 amino acid peptide called amyloid beta whose abnormally folded fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients. Gamma secretase is also critical in the related processing of several other type I integral membrane proteins, such as Notch, ErbB4, E-cadherin, N-cadherin, ephrin-B2, or CD44.

Presenilins are a family of related multi-pass transmembrane proteins which constitute the catalytic subunits of the gamma-secretase intramembrane protease protein complex. They were first identified in screens for mutations causing early onset forms of familial Alzheimer's disease by Peter St George-Hyslop. Vertebrates have two presenilin genes, called PSEN1 that codes for presenilin 1 (PS-1) and PSEN2 that codes for presenilin 2 (PS-2). Both genes show conservation between species, with little difference between rat and human presenilins. The nematode worm C. elegans has two genes that resemble the presenilins and appear to be functionally similar, sel-12 and hop-1.

A Disintegrin and metalloproteinase domain-containing protein 10, also known as ADAM10 or CDw156 or CD156c is a protein that in humans is encoded by the ADAM10 gene.

Kallikrein-6 is a protein that in humans is encoded by the KLK6 gene. Kallikrein-6 is also referred to as neurosin, protease M, hK6, or zyme. It is a 223 amino acid sequence, derived from its 244 original form, which contains a 16 residue presignal and 5 residue activation peptide.

Pitrilysin metallopeptidase 1 also known as presequence protease, mitochondrial (PreP) and metalloprotease 1 (MTP-1) is an enzyme that in humans is encoded by the PITRM1 gene. It is also sometimes called metalloprotease 1 (MP1).PreP facilitates proteostasis by utilizing an ~13300-A(3) catalytic chamber to degrade toxic peptides, including mitochondrial presequences and β-amyloid. Deficiency of PreP is found associated with Alzheimer's disease. Reduced levels of PreP via RNAi mediated knockdown have been shown to lead to defective maturation of the protein Frataxin.
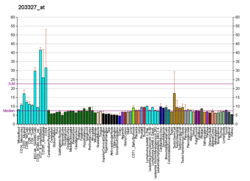
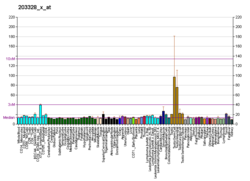
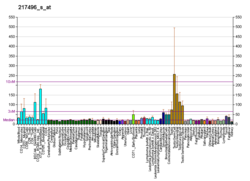
The biomarkers of Alzheimer's disease are neurochemical indicators used to assess the risk or presence of the disease. The biomarkers can be used to diagnose Alzheimer's disease (AD) in a very early stage, but they also provide objective and reliable measures of disease progress. It is imperative to diagnose AD disease as soon as possible, because neuropathologic changes of AD precede the symptoms by years. It is well known that amyloid beta (Aβ) is a good indicator of AD disease, which has facilitated doctors to accurately pre-diagnose cases of AD. When Aβ peptide is released by proteolytic cleavage of amyloid-beta precursor protein, some Aβ peptides that are solubilized are detected in CSF and blood plasma which makes AB peptides a promising candidate for biological markers. It has been shown that the amyloid beta biomarker shows 80% or above sensitivity and specificity, in distinguishing AD from dementia. It is believed that amyloid beta as a biomarker will provide a future for diagnosis of AD and eventually treatment of AD.
Intramembrane proteases (IMPs), also known as intramembrane-cleaving proteases (I-CLiPs), are enzymes that have the property of cleaving transmembrane domains of integral membrane proteins. All known intramembrane proteases are themselves integral membrane proteins with multiple transmembrane domains, and they have their active sites buried within the lipid bilayer of cellular membranes. Intramembrane proteases are responsible for proteolytic cleavage in the cell signaling process known as regulated intramembrane proteolysis (RIP).

Phospholipase D3, also known as PLD3, is a protein that in humans is encoded by the PLD3 gene. PLD3 belongs to the phospholipase D superfamily because it contains the two HKD motifs common to members of the phospholipase D family, however, it has no known catalytic function similar to PLD1 or PLD2. PLD3 serves as a ssDNA 5' exonuclease in antigen presenting cells. PLD3 is highly expressed in the brain in both humans and mice, and is mainly localized in the endoplasmic reticulum (ER) and the lysosome.
The ion channel hypothesis of Alzheimer's disease (AD), also known as the channel hypothesis or the amyloid beta ion channel hypothesis, is a more recent variant of the amyloid hypothesis of AD, which identifies amyloid beta (Aβ) as the underlying cause of neurotoxicity seen in AD. While the traditional formulation of the amyloid hypothesis pinpoints insoluble, fibrillar aggregates of Aβ as the basis of disruption of calcium ion homeostasis and subsequent apoptosis in AD, the ion channel hypothesis in 1993 introduced the possibility of an ion-channel-forming oligomer of soluble, non-fibrillar Aβ as the cytotoxic species allowing unregulated calcium influx into neurons in AD.