A biological membrane, biomembrane or cell membrane is a selectively permeable membrane that separates the interior of a cell from the external environment or creates intracellular compartments by serving as a boundary between one part of the cell and another. Biological membranes, in the form of eukaryotic cell membranes, consist of a phospholipid bilayer with embedded, integral and peripheral proteins used in communication and transportation of chemicals and ions. The bulk of lipids in a cell membrane provides a fluid matrix for proteins to rotate and laterally diffuse for physiological functioning. Proteins are adapted to high membrane fluidity environment of the lipid bilayer with the presence of an annular lipid shell, consisting of lipid molecules bound tightly to the surface of integral membrane proteins. The cell membranes are different from the isolating tissues formed by layers of cells, such as mucous membranes, basement membranes, and serous membranes.

The endomembrane system is composed of the different membranes (endomembranes) that are suspended in the cytoplasm within a eukaryotic cell. These membranes divide the cell into functional and structural compartments, or organelles. In eukaryotes the organelles of the endomembrane system include: the nuclear membrane, the endoplasmic reticulum, the Golgi apparatus, lysosomes, vesicles, endosomes, and plasma (cell) membrane among others. The system is defined more accurately as the set of membranes that forms a single functional and developmental unit, either being connected directly, or exchanging material through vesicle transport. Importantly, the endomembrane system does not include the membranes of plastids or mitochondria, but might have evolved partially from the actions of the latter.

Facilitated diffusion is the process of spontaneous passive transport of molecules or ions across a biological membrane via specific transmembrane integral proteins. Being passive, facilitated transport does not directly require chemical energy from ATP hydrolysis in the transport step itself; rather, molecules and ions move down their concentration gradient reflecting its diffusive nature.
Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs may consist of one or several alpha-helices or a transmembrane beta barrel. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in size and hydrophobicity; they may adopt organelle-specific properties.

The lipid bilayer is a thin polar membrane made of two layers of lipid molecules. These membranes are flat sheets that form a continuous barrier around all cells. The cell membranes of almost all organisms and many viruses are made of a lipid bilayer, as are the nuclear membrane surrounding the cell nucleus, and membranes of the membrane-bound organelles in the cell. The lipid bilayer is the barrier that keeps ions, proteins and other molecules where they are needed and prevents them from diffusing into areas where they should not be. Lipid bilayers are ideally suited to this role, even though they are only a few nanometers in width, because they are impermeable to most water-soluble (hydrophilic) molecules. Bilayers are particularly impermeable to ions, which allows cells to regulate salt concentrations and pH by transporting ions across their membranes using proteins called ion pumps.

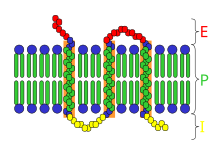
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane. Peripheral membrane proteins are transiently associated with the cell membrane.

A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.

Peripheral membrane proteins, or extrinsic membrane proteins, are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.

Biochemistry is the study of the chemical processes in living organisms. It deals with the structure and function of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules.
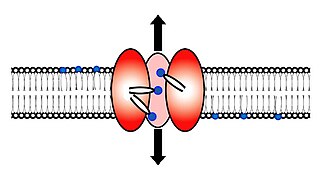
Scramblase is a protein responsible for the translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane. In humans, phospholipid scramblases (PLSCRs) constitute a family of five homologous proteins that are named as hPLSCR1–hPLSCR5. Scramblases are members of the general family of transmembrane lipid transporters known as flippases. Scramblases are distinct from flippases and floppases. Scramblases, flippases, and floppases are three different types of enzymatic groups of phospholipid transportation enzymes. The inner-leaflet, facing the inside of the cell, contains negatively charged amino-phospholipids and phosphatidylethanolamine. The outer-leaflet, facing the outside environment, contains phosphatidylcholine and sphingomyelin. Scramblase is an enzyme, present in the cell membrane, that can transport (scramble) the negatively charged phospholipids from the inner-leaflet to the outer-leaflet, and vice versa.

Flippases are transmembrane lipid transporter proteins located in the membrane which belong to ABC transporter or P4-type ATPase families. They are responsible for aiding the movement of phospholipid molecules between the two leaflets that compose a cell's membrane. This is necessary to continue their normal function of growth and mobility. The possibility of active maintenance of an asymmetric distribution of molecules in the phospholipid bilayer was predicted in the early 1970s by Mark Bretscher. Although phospholipids diffuse rapidly in the plane of the membrane, their polar head groups cannot pass easily through the hydrophobic center of the bilayer, limiting their diffusion in this dimension. Some flippases - often instead called scramblases - are energy-independent and bidirectional, causing reversible equilibration of phospholipid between the two sides of the membrane, whereas others are energy-dependent and unidirectional, using energy from ATP hydrolysis to pump the phospholipid in a preferred direction. Flippases are described as transporters that move lipids from the exoplasmic to the cytosolic face, while floppases transport in the reverse direction.
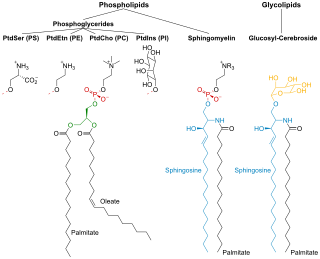
Membrane lipids are a group of compounds which form the lipid bilayer of the cell membrane. The three major classes of membrane lipids are phospholipids, glycolipids, and cholesterol. Lipids are amphiphilic: they have one end that is soluble in water ('polar') and an ending that is soluble in fat ('nonpolar'). By forming a double layer with the polar ends pointing outwards and the nonpolar ends pointing inwards membrane lipids can form a 'lipid bilayer' which keeps the watery interior of the cell separate from the watery exterior. The arrangements of lipids and various proteins, acting as receptors and channel pores in the membrane, control the entry and exit of other molecules and ions as part of the cell's metabolism. In order to perform physiological functions, membrane proteins are facilitated to rotate and diffuse laterally in two dimensional expanse of lipid bilayer by the presence of a shell of lipids closely attached to protein surface, called annular lipid shell.
A model lipid bilayer is any bilayer assembled in vitro, as opposed to the bilayer of natural cell membranes or covering various sub-cellular structures like the nucleus. They are used to study the fundamental properties of biological membranes in a simplified and well-controlled environment, and increasingly in bottom-up synthetic biology for the construction of artificial cells. A model bilayer can be made with either synthetic or natural lipids. The simplest model systems contain only a single pure synthetic lipid. More physiologically relevant model bilayers can be made with mixtures of several synthetic or natural lipids.
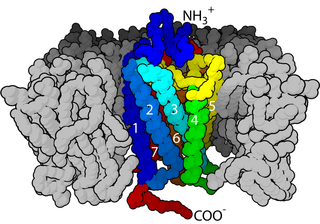
Cell surface receptors are receptors that are embedded in the plasma membrane of cells. They act in cell signaling by receiving extracellular molecules. They are specialized integral membrane proteins that allow communication between the cell and the extracellular space. The extracellular molecules may be hormones, neurotransmitters, cytokines, growth factors, cell adhesion molecules, or nutrients; they react with the receptor to induce changes in the metabolism and activity of a cell. In the process of signal transduction, ligand binding affects a cascading chemical change through the cell membrane.
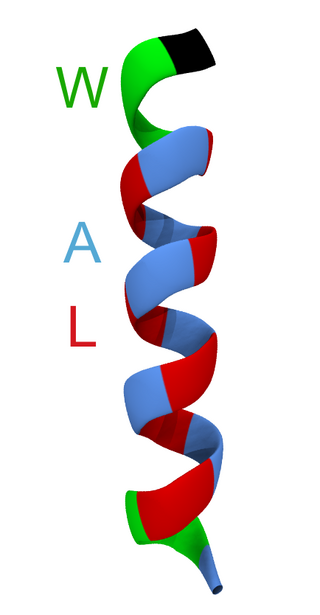
WALP peptides are a class of synthesized, membrane-spanning α-helices composed of tryptophan (W), alanine (A), and leucine (L) amino acids. They are designed to study properties of proteins in lipid membranes such as orientation, extent of insertion, and hydrophobic mismatch.
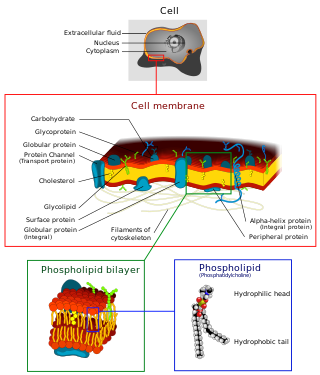
The cell membrane is a biological membrane that separates and protects the interior of a cell from the outside environment. The cell membrane consists of a lipid bilayer, made up of two layers of phospholipids with cholesterols interspersed between them, maintaining appropriate membrane fluidity at various temperatures. The membrane also contains membrane proteins, including integral proteins that span the membrane and serve as membrane transporters, and peripheral proteins that loosely attach to the outer (peripheral) side of the cell membrane, acting as enzymes to facilitate interaction with the cell's environment. Glycolipids embedded in the outer lipid layer serve a similar purpose. The cell membrane controls the movement of substances in and out of a cell, being selectively permeable to ions and organic molecules. In addition, cell membranes are involved in a variety of cellular processes such as cell adhesion, ion conductivity, and cell signalling and serve as the attachment surface for several extracellular structures, including the cell wall and the carbohydrate layer called the glycocalyx, as well as the intracellular network of protein fibers called the cytoskeleton. In the field of synthetic biology, cell membranes can be artificially reassembled.
This is a list of articles on biophysics.

Collagen α-1 (XXIII) chain is a protein encoded by COL23A1 gene, which is located on chromosome 5q35 in humans, and on chromosome 11B1+2 in mice. The location of this gene was discovered by genomic sequence analysis.

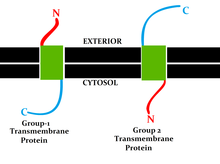
A single-pass membrane protein also known as single-spanning protein or bitopic protein is a transmembrane protein that spans the lipid bilayer only once. These proteins may constitute up to 50% of all transmembrane proteins, depending on the organism, and contribute significantly to the network of interactions between different proteins in cells, including interactions via transmembrane alpha helices. They usually include one or several water-soluble domains situated at the different sides of biological membranes, for example in single-pass transmembrane receptors. Some of them are small and serve as regulatory or structure-stabilizing subunits in large multi-protein transmembrane complexes, such as photosystems or the respiratory chain. A 2013 estimate identified about 1300 single-pass membrane proteins in the human genome.