
Staphylococcus aureus is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Methicillin-resistant Staphylococcus aureus (MRSA) is a group of gram-positive bacteria that are genetically distinct from other strains of Staphylococcus aureus. MRSA is responsible for several difficult-to-treat infections in humans. It caused more than 100,000 deaths worldwide attributable to antimicrobial resistance in 2019.

Klebsiella pneumoniae is a Gram-negative, non-motile, encapsulated, lactose-fermenting, facultative anaerobic, rod-shaped bacterium. It appears as a mucoid lactose fermenter on MacConkey agar.

Vancomycin-resistant Staphylococcus aureus (VRSA) are strains of Staphylococcus aureus that have acquired resistance to the glycopeptide antibiotic vancomycin. Bacteria can acquire resistant genes either by random mutation or through the transfer of DNA from one bacterium to another. Resistance genes interfere with the normal antibiotic function and allow a bacteria to grow in the presence of the antibiotic. Resistance in VRSA is conferred by the plasmid-mediated vanA gene and operon. Although VRSA infections are uncommon, VRSA is often resistant to other types of antibiotics and a potential threat to public health because treatment options are limited. VRSA is resistant to many of the standard drugs used to treat S. aureus infections. Furthermore, resistance can be transferred from one bacterium to another.

Stanley "Stan" Falkow was an American microbiologist and a professor of microbiology at Georgetown University, University of Washington, and Stanford University School of Medicine. Falkow is known as the father of the field of molecular microbial pathogenesis. He formulated molecular Koch's postulates, which have guided the study of the microbial determinants of infectious diseases since the late 1980s. Falkow spent over 50 years uncovering molecular mechanisms of how bacteria cause disease and how to disarm them. Falkow also was one of the first scientists to investigate antimicrobial resistance, and presented his research extensively to scientific, government, and lay audiences explaining the spread of resistance from one organism to another, now known as horizontal gene transfer, and the implications of this phenomenon on our ability to combat infections in the future.

Staphylococcus epidermidis is a Gram-positive bacterium, and one of over 40 species belonging to the genus Staphylococcus. It is part of the normal human microbiota, typically the skin microbiota, and less commonly the mucosal microbiota and also found in marine sponges. It is a facultative anaerobic bacteria. Although S. epidermidis is not usually pathogenic, patients with compromised immune systems are at risk of developing infection. These infections are generally hospital-acquired. S. epidermidis is a particular concern for people with catheters or other surgical implants because it is known to form biofilms that grow on these devices. Being part of the normal skin microbiota, S. epidermidis is a frequent contaminant of specimens sent to the diagnostic laboratory.

Panton–Valentine leukocidin (PVL) is a cytotoxin—one of the β-pore-forming toxins. The presence of PVL is associated with increased virulence of certain strains (isolates) of Staphylococcus aureus. It is present in the majority of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) isolates studied and is the cause of necrotic lesions involving the skin or mucosa, including necrotic hemorrhagic pneumonia. PVL creates pores in the membranes of infected cells. PVL is produced from the genetic material of a bacteriophage that infects Staphylococcus aureus, making it more virulent.
Lysostaphin is a Staphylococcus simulans metalloendopeptidase. It can function as a bacteriocin (antimicrobial) against Staphylococcus aureus.
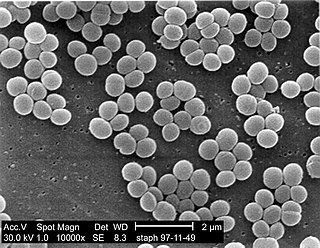
A staphylococcal infection or staph infection is an infection caused by members of the Staphylococcus genus of bacteria.
mecA is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics.

Staphylococcus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical (cocci), and form in grape-like clusters. Staphylococcus species are facultative anaerobic organisms.
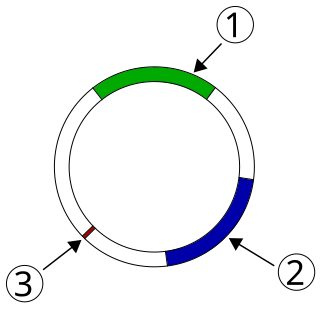
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.

Staphylococcus hyicus is a Gram-positive, facultatively anaerobic bacterium in the genus Staphylococcus. It consists of clustered cocci and forms white circular colonies when grown on blood agar. S. hyicus is a known animal pathogen. It causes disease in poultry, cattle, horses, and pigs. Most notably, it is the agent that causes porcine exudative epidermitis, also known as greasy pig disease, in piglets. S. hyicus is generally considered to not be zoonotic, however it has been shown to be able to cause bacteremia and sepsis in humans.
Carbapenem-resistant Enterobacteriaceae (CRE) or carbapenemase-producing Enterobacteriaceae (CPE) are Gram-negative bacteria that are resistant to the carbapenem class of antibiotics, considered the drugs of last resort for such infections. They are resistant because they produce an enzyme called a carbapenemase that disables the drug molecule. The resistance can vary from moderate to severe. Enterobacteriaceae are common commensals and infectious agents. Experts fear CRE as the new "superbug". The bacteria can kill up to half of patients who get bloodstream infections. Tom Frieden, former head of the Centers for Disease Control and Prevention has referred to CRE as "nightmare bacteria". Examples of enzymes found in certain types of CRE are KPC and NDM. KPC and NDM are enzymes that break down carbapenems and make them ineffective. Both of these enzymes, as well as the enzyme VIM have also been reported in Pseudomonas.
Staphylococcus schleiferi is a Gram-positive, cocci-shaped bacterium of the family Staphylococcaceae. It is facultatively anaerobic, coagulase-variable, and can be readily cultured on blood agar where the bacterium tends to form opaque, non-pigmented colonies and beta (β) hemolysis. There exists two subspecies under the species S. schleiferi: Staphylococcus schleiferi subsp. schleiferi and Staphylococcus schleiferi subsp. coagulans.

Phyllis Margaret Rountree was an Australian microbiologist and bacteriologist. She was an expert in staphylococcal infections.

Georg Peters was a German physician, microbiologist and university professor. From 1992 until his fatal mountain accident he headed the Institute of Medical Microbiology at the University of Münster. He was an internationally recognised expert in the field of staphylococci and the infectious diseases caused by them, to which he had devoted himself since the beginning of his scientific career.
Kerry L. LaPlante is an American pharmacist, academic and researcher. She is a Professor of Pharmacy and the Chair of the Department of Pharmacy Practice at the University of Rhode Island, an Adjunct Professor of Medicine at Brown University, an Infectious Diseases Pharmacotherapy Specialist, and the Director of the Rhode Island Infectious Diseases Fellowship and Research Programs at the Veterans Affairs Medical Center in Providence, Rhode Island.
Vincent A. Fischetti is a world renowned American microbiologist and immunologist. He is Professor of and Head of the Laboratory of Bacterial Pathogenesis and Immunology at Rockefeller University in New York City. His Laboratory is the oldest continuous laboratory at Rockefeller that started in 1926 and headed by 4 leading scientists over its near 100 year history: Homer Swift, Maclyn McCarty, Emil Gotschlich and now Vincent Fischetti. Keeping with the historical theme of infectious diseases, Fischetti's primary areas of research are bacterial pathogenesis, bacterial genomics, immunology, virology, microbiology, and therapeutics. He was the first scientist to clone and sequence a surface protein on gram-positive bacteria, the M protein from S. pyogenes, and determine its unique coiled-coil structure. He also was the first use phage lysins as a therapeutic and an effective alternative to conventional antibiotics.
Richard P. Novick is an American microbiologist best known for his work in the fields of plasmid biology, staphylococcal pathobiology and antimicrobial resistance. He is the Recanati Family Professor of Science, Emeritus, at NYU Grossman School of Medicine and is a member of the American National Academy of Sciences. Novick has published over 250 peer-reviewed articles, and several book reviews for the Times Literary Supplement, and is a member of the Editorial Board of the Proceedings of the National Academy of Sciences.












