Related Research Articles

Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.
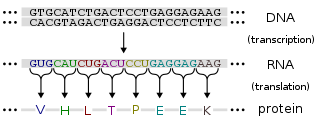
Protein production is the biotechnological process of generating a specific protein. It is typically achieved by the manipulation of gene expression in an organism such that it expresses large amounts of a recombinant gene. This includes the transcription of the recombinant DNA to messenger RNA (mRNA), the translation of mRNA into polypeptide chains, which are ultimately folded into functional proteins and may be targeted to specific subcellular or extracellular locations.
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be co-transcribed to define an operon.

The lactose operon is an operon required for the transport and metabolism of lactose in E. coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.
A transcriptional activator is a protein that increases transcription of a gene or set of genes. Activators are considered to have positive control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain".
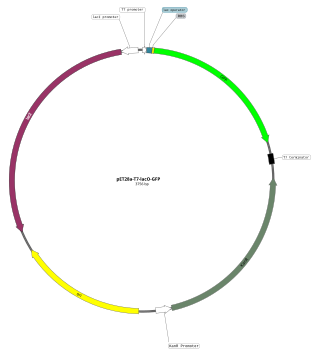
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector is used to introduce a specific gene into a target cell, and can commandeer the cell's mechanism for protein synthesis to produce the protein encoded by the gene. Expression vectors are the basic tools in biotechnology for the production of proteins.

Bacteriophage T7 is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
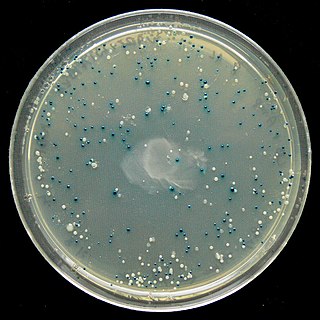
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used. DNA of transformation is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.
The L-arabinose operon, also called the ara or araBAD operon, is an operon required for the breakdown of the five-carbon sugar L-arabinose in Escherichia coli. The L-arabinose operon contains three structural genes: araB, araA, araD, which encode for three metabolic enzymes that are required for the metabolism of L-arabinose. AraB (ribulokinase), AraA, AraD produced by these genes catalyse conversion of L-arabinose to an intermediate of the pentose phosphate pathway, D-xylulose-5-phosphate.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

Spot 42 (spf) RNA is a regulatory non-coding bacterial small RNA encoded by the spf gene. Spf is found in gammaproteobacteria and the majority of experimental work on Spot42 has been performed in Escherichia coli and recently in Aliivibrio salmonicida. In the cell Spot42 plays essential roles as a regulator in carbohydrate metabolism and uptake, and its expression is activated by glucose, and inhibited by the cAMP-CRP complex.
In molecular cloning, a vector is any particle used as a vehicle to artificially carry a foreign nucleic sequence – usually DNA – into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors have an origin of replication, a multicloning site, and a selectable marker.
The gal operon is a prokaryotic operon, which encodes enzymes necessary for galactose metabolism. Repression of gene expression for this operon works via binding of repressor molecules to two operators. These repressors dimerize, creating a loop in the DNA. The loop as well as hindrance from the external operator prevent RNA polymerase from binding to the promoter, and thus prevent transcription. Additionally, since the metabolism of galactose in the cell is involved in both anabolic and catabolic pathways, a novel regulatory system using two promoters for differential repression has been identified and characterized within the context of the gal operon.

Bacteriophage P2, scientific name Escherichia virus P2, is a temperate phage that infects E. coli. It is a tailed virus with a contractile sheath and is thus classified in the genus Peduovirus, subfamily Peduovirinae, family Myoviridae within order Caudovirales. This genus of viruses includes many P2-like phages as well as the satellite phage P4.

Daisy Roulland-Dussoix was a Swiss molecular microbiologist. She was one of the discoverers of restriction enzymes during her doctoral studies. There is controversy over whether she should have received the 1978 Nobel prize in Physiology and Medicine, which was awarded to Hamilton O. Smith, Daniel Nathans, and Werner Arber.
SCAR-less genome editing Scarless Cas9 Assisted Recombineering (no-SCAR) is an editing method that is able to manipulate the Escherichia coli genome. The system relies on recombineering whereby DNA sequences are combined and manipulated through homologous recombination. No-SCAR is able to manipulate the E. coli genome without the use of the chromosomal markers detailed in previous recombineering methods. Instead, in this method, the λ-Red recombination system facilitates donor DNA integration while Cas9 cleaves double-stranded DNA to counter-select against wild-type cells. Although λ-Red and Cas9 genome editing are widely used technologies, the no-SCAR method is novel in combining the two functions; this technique is able to establish point mutations, gene deletions, and short sequence insertions in several genomic loci with increased efficiency and time sensitivity.
Transcription-translation coupling is a mechanism of gene expression regulation in which synthesis of an mRNA (transcription) is affected by its concurrent decoding (translation). In prokaryotes, mRNAs are translated while they are transcribed. This allows communication between RNA polymerase, the multisubunit enzyme that catalyzes transcription, and the ribosome, which catalyzes translation. Coupling involves both direct physical interactions between RNA polymerase and the ribosome, as well as ribosome-induced changes to the structure and accessibility of the intervening mRNA that affect transcription.
The T7 expression system is used in the field of microbiology to clone recombinant DNA using strains of E. coli. It is the most popular system for expressing recombinant proteins in E. coli.
References
- 1 2 Studier, FW; Moffatt, BA (5 May 1986). "Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes". Journal of Molecular Biology. 189 (1): 113–30. doi:10.1016/0022-2836(86)90385-2. PMID 3537305.
- 1 2 Noel RJ, Reznikoff WS (10 March 2000). "Structural Studies of lacUV5-RNA Polymerase Interactions in Vitro". Journal of Biological Chemistry. 275 (11): 7708–7712. doi: 10.1074/jbc.275.11.7708 . PMID 10713082.
- ↑ Pribnow, D (1975). "Bacteriophage T7 Early Promoters: Nucleotide Sequences of Two RNA Polymerase Binding Sites". Journal of Molecular Biology. 99 (3): 419–443. doi:10.1016/S0022-2836(75)80136-7. PMID 765476.
- ↑ Silverstone, AE; Arditti, RR; Magasanik, B (July 1970). "Catabolite-insensitive revertants of lac promoter mutants". Proceedings of the National Academy of Sciences of the United States of America. 66 (3): 773–9. Bibcode:1970PNAS...66..773S. doi: 10.1073/pnas.66.3.773 . PMC 283117 . PMID 4913210.
- ↑ Fuller, F (1982). "A family of cloning vectors containing the lacUV5 promoter". Gene. 19 (1): 43–54. doi:10.1016/0378-1119(82)90187-1. PMID 6292048.
- ↑ "Escherichia coli strain BLR(DE3) chromosome, complete genome". GenBank. 2017-04-13. Retrieved 21 April 2019.
- ↑ "Escherichia coli strain BLR(DE3) chromosome, LacZ". GenBank. Retrieved 21 April 2019.