Related Research Articles

Bioinformatics is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for in silico analyses of biological queries using computational and statistical techniques.

Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer engineering which uses bioengineering to build computers.
The Open Bioinformatics Foundation is a non-profit, volunteer-run organization focused on supporting open source programming in bioinformatics. The mission of the foundation is to support the development of open source toolkits for bioinformatics, organise developer-centric hackathon events and generally assist in the development and promotion of open source software development in the life sciences. The foundation also organises and runs the annual Bioinformatics Open Source Conference, a satellite meeting of the Intelligent Systems for Molecular Biology conference. The foundation participates in the Google Summer of Code, acting as an umbrella organisation for individual bioinformatics-related projects.
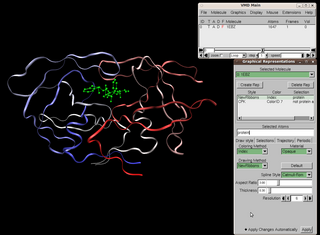
Visual Molecular Dynamics (VMD) is a molecular modelling and visualization computer program. VMD is developed mainly as a tool to view and analyze the results of molecular dynamics simulations. It also includes tools for working with volumetric data, sequence data, and arbitrary graphics objects. Molecular scenes can be exported to external rendering tools such as POV-Ray, RenderMan, Tachyon, Virtual Reality Modeling Language (VRML), and many others. Users can run their own Tcl and Python scripts within VMD as it includes embedded Tcl and Python interpreters. VMD runs on Unix, Apple Mac macOS, and Microsoft Windows. VMD is available to non-commercial users under a distribution-specific license which permits both use of the program and modification of its source code, at no charge.
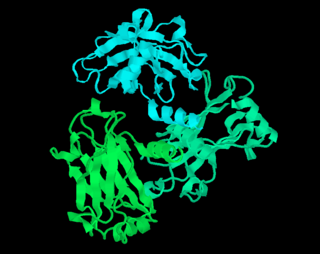
RasMol is a computer program written for molecular graphics visualization intended and used mainly to depict and explore biological macromolecule structures, such as those found in the Protein Data Bank. It was originally developed by Roger Sayle in the early 1990s.

PyMOL is an open source but proprietary molecular visualization system created by Warren Lyford DeLano. It was commercialized initially by DeLano Scientific LLC, which was a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities. It is currently commercialized by Schrödinger, Inc. As the original software license was a permissive licence, they were able to remove it; new versions are no longer released under the Python license, but under a custom license, and some of the source code is no longer released. PyMOL can produce high-quality 3D images of small molecules and biological macromolecules, such as proteins. According to the original author, by 2009, almost a quarter of all published images of 3D protein structures in the scientific literature were made using PyMOL.
Bioconductor is a free, open source and open development software project for the analysis and comprehension of genomic data generated by wet lab experiments in molecular biology.

Jmol is computer software for molecular modelling chemical structures in 3-dimensions. Jmol returns a 3D representation of a molecule that may be used as a teaching tool, or for research e.g., in chemistry and biochemistry. It is written in the programming language Java, so it can run on the operating systems Windows, macOS, Linux, and Unix, if Java is installed. It is free and open-source software released under a GNU Lesser General Public License (LGPL) version 2.0. A standalone application and a software development kit (SDK) exist that can be integrated into other Java applications, such as Bioclipse and Taverna.

The Chemistry Development Kit (CDK) is computer software, a library in the programming language Java, for chemoinformatics and bioinformatics. It is available for Windows, Linux, Unix, and macOS. It is free and open-source software distributed under the GNU Lesser General Public License (LGPL) 2.0.

BALL is a C++ class framework and set of algorithms and data structures for molecular modelling and computational structural bioinformatics, a Python interface to this library, and a graphical user interface to BALL, the molecule viewer BALLView.
Physiomics is a systematic study of physiome in biology. Physiomics employs bioinformatics to construct networks of physiological features that are associated with genes, proteins and their networks. A few of the methods for determining individual relationships between the DNA sequence and physiological function include metabolic pathway engineering and RNAi analysis. The relationships derived from methods such as these are organized and processed computationally to form distinct networks. Computer models use these experimentally determined networks to develop further predictions of gene function.

Avogadro is a molecule editor and visualizer designed for cross-platform use in computational chemistry, molecular modeling, bioinformatics, materials science, and related areas. It is extensible via a plugin architecture.
Biology data visualization is a branch of bioinformatics concerned with the application of computer graphics, scientific visualization, and information visualization to different areas of the life sciences. This includes visualization of sequences, genomes, alignments, phylogenies, macromolecular structures, systems biology, microscopy, and magnetic resonance imaging data. Software tools used for visualizing biological data range from simple, standalone programs to complex, integrated systems.
geWorkbench is an open-source software platform for integrated genomic data analysis. It is a desktop application written in the programming language Java. geWorkbench uses a component architecture. As of 2016, there are more than 70 plug-ins available, providing for the visualization and analysis of gene expression, sequence, and structure data.
Pharmaceutical bioinformatics is a research field related to bioinformatics but with the focus on studying biological and chemical processes in the pharmaceutical area; to understand how xenobiotics interact with the human body and the drug discovery process.
References
- ↑ Biopython License
- ↑ "GeWorkbench License". geWorkbench. Columbia University. 15 June 2014.