The miR-9 microRNA, is a short non-coding RNA gene involved in gene regulation. The mature ~21nt miRNAs are processed from hairpin precursor sequences by the Dicer enzyme. The dominant mature miRNA sequence is processed from the 5' arm of the mir-9 precursor, and from the 3' arm of the mir-79 precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In vertebrates, miR-9 is highly expressed in the brain, and is suggested to regulate neuronal differentiation. A number of specific targets of miR-9 have been proposed, including the transcription factor REST and its partner CoREST.

The miR-124 microRNA precursor is a small non-coding RNA molecule that has been identified in flies, nematode worms, mouse and human. The mature ~21 nucleotide microRNAs are processed from hairpin precursor sequences by the Dicer enzyme, and in this case originates from the 3' arm. miR-124 has been found to be the most abundant microRNA expressed in neuronal cells. Experiments to alter expression of miR-124 in neural cells did not appear to affect differentiation. However these results are controversial since other reports have described a role for miR-124 during neuronal differentiation.

The miR-129 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. This microRNA was first experimentally characterised in mouse and homologues have since been discovered in several other species, such as humans, rats and zebrafish. The mature sequence is excised by the Dicer enzyme from the 5' arm of the hairpin. It was elucidated by Calin et al. that miR-129-1 is located in a fragile site region of the human genome near a specific site, FRA7H in chromosome 7q32, which is a site commonly deleted in many cancers. miR-129-2 is located in 11p11.2.

The miR-17 microRNA precursor family are a group of related small non-coding RNA genes called microRNAs that regulate gene expression. The microRNA precursor miR-17 family, includes miR-20a/b, miR-93, and miR-106a/b. With the exception of miR-93, these microRNAs are produced from several microRNA gene clusters, which apparently arose from a series of ancient evolutionary genetic duplication events, and also include members of the miR-19, and miR-25 families. These clusters are transcribed as long non-coding RNA transcripts that are processed to form ~70 nucleotide microRNA precursors, that are subsequently processed by the Dicer enzyme to give a ~22 nucleotide products. The mature microRNA products are thought to regulate expression levels of other genes through complementarity to the 3' UTR of specific target messenger RNA.

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, and these are called miR-1-1 and miR-1-2.
The miR-34 microRNA precursor family are non-coding RNA molecules that, in mammals, give rise to three major mature miRNAs. The miR-34 family members were discovered computationally and later verified experimentally. The precursor miRNA stem-loop is processed in the cytoplasm of the cell, with the predominant miR-34 mature sequence excised from the 5' arm of the hairpin.

microRNA 21 also known as hsa-mir-21 or miRNA21 is a mammalian microRNA that is encoded by the MIR21 gene.
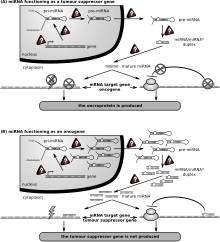
An oncomir is a microRNA (miRNA) that is associated with cancer. MicroRNAs are short RNA molecules about 22 nucleotides in length. Essentially, miRNAs specifically target certain messenger RNAs (mRNAs) to prevent them from coding for a specific protein. The dysregulation of certain microRNAs (oncomirs) has been associated with specific cancer forming (oncogenic) events. Many different oncomirs have been identified in numerous types of human cancers.

MiR-155 is a microRNA that in humans is encoded by the MIR155 host gene or MIR155HG. MiR-155 plays a role in various physiological and pathological processes. Exogenous molecular control in vivo of miR-155 expression may inhibit malignant growth, viral infections, and enhance the progression of cardiovascular diseases.

In molecular biology mir-126 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several pre- and post-transcription mechanisms.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.

In molecular biology, mir-145 microRNA is a short RNA molecule that in humans is encoded by the MIR145 gene. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology, the miR-200 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by binding and cleaving mRNAs or inhibiting translation. The miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. There is growing evidence to suggest that miR-200 microRNAs are involved in cancer metastasis.

In molecular biology MicroRNA-223 (miR-223) is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. miR-223 is a hematopoietic specific microRNA with crucial functions in myeloid lineage development. It plays an essential role in promoting granulocytic differentiation while also being associated with the suppression of erythrocytic differentiation. miR-223 is commonly repressed in hepatocellular carcinoma and leukemia. Higher expression levels of miRNA-223 are associated with extranodal marginal-zone lymphoma of mucosa-associated lymphoid tissue of the stomach and recurrent ovarian cancer. In some cancers the microRNA-223 down-regulation is correlated with higher tumor burden, disease aggressiveness, and poor prognostic factors. MicroRNA-223 is also associated with rheumatoid arthritis, sepsis, type 2 diabetes, and hepatic ischemia.
miR-31 has been characterised as a tumour suppressor miRNA, with its levels varying in breast cancer cells according to the metastatic state of the tumour. From its typical abundance in healthy tissue is a moderate decrease in non-metastatic breast cancer cell lines, and levels are almost completely absent in mouse and human metastatic breast cancer cell lines. Mir-31-5p has also been observed upregulated in Zinc Deficient rats compared to normal in ESCC and in other types of cancers when using this animal model. There has also been observed a strong encapsulation of tumour cells expressing miR-31, as well as a reduced cell survival rate. miR-31's antimetastatic effects therefore make it a potential therapeutic target for breast cancer. However, these two papers were formally retracted by the authors in 2015.
In molecular biology mir-326 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-497 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-885 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
MicroRNA-125 (miR-125) is a highly conserved microRNA family consisting of miR-125a and miR-125b. MiR-125 can be found throughout diverse species from nematode to humans. MiR-125 family members are involved in cell differentiation, proliferation and apoptosis as a result of targeting messenger RNAs related to these cellular processes. By affecting these cellular processes, miR-125 can cause promotion or suppression of pathological processes including carcinogenesis, muscle abnormalities, neurological disorders and pathologies of the immune system. Moreover, miR-125 also plays an important role in normal immune functions and was described to affect development and function of immune cells as well as playing role in immunological host defense in response to bacterial and viral infections.