
A microRNA is a small single-stranded non-coding RNA molecule found in plants, animals and some viruses, that functions in RNA silencing and post-transcriptional regulation of gene expression. miRNAs function via base-pairing with complementary sequences within mRNA molecules. As a result, these mRNA molecules are silenced, by one or more of the following processes: (1) cleavage of the mRNA strand into two pieces, (2) destabilization of the mRNA through shortening of its poly(A) tail, and (3) less efficient translation of the mRNA into proteins by ribosomes.

Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the DICER1 gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA).
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).
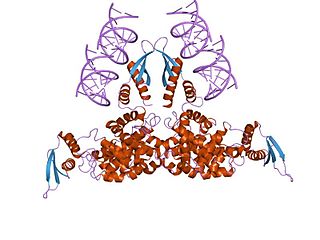
Ribonuclease III (BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.
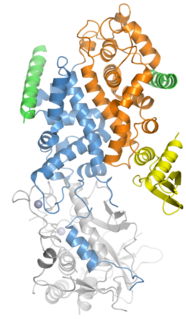
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the DROSHA gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosis and HIV-1 replication.

The microprocessor complex subunit DGCR8(DiGeorge syndrome critical region 8) is a protein that in humans is encoded by the DGCR8 gene. In other animals, particularly the common model organisms Drosophila melanogaster and Caenorhabditis elegans, the protein is known as Pasha. It is a required component of the RNA interference pathway.
Mirtrons are a type of microRNAs that are located in the introns of the mRNA encoding host genes. These short hairpin introns formed via atypical miRNA biogenesis pathways. Mirtrons arise from the spliced-out introns and are known to function in gene expression.

RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
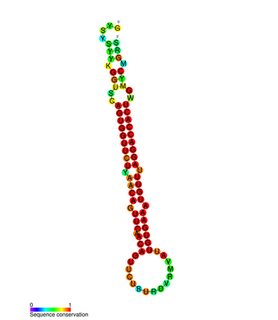
In molecular biology miR-203 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms, such as translational repression and Argonaute-catalyzed messenger RNA cleavage. miR-203 has been identified as a skin-specific microRNA, and it forms an expression gradient that defines the boundary between proliferative epidermal basal progenitors and terminally differentiating suprabasal cells. It has also been found upregulated in psoriasis and differentially expressed in some types of cancer.

V. Narry Kim is a South Korean biochemist and microbiologist, best known for her work on microRNA biogenesis. Her pioneering studies have laid the groundwork for the biology of microRNA and contributed to the improvement of RNA interference technologies.
In molecular biology, small nucleolar RNA derived microRNAs are microRNAs (miRNA) derived from small nucleolar RNA (snoRNA). MicroRNAs are usually derived from precursors known as pre-miRNAs, these pre-miRNAs are recognised and cleaved from a pri-miRNA precursor by the Pasha and Drosha proteins. However some microRNAs, mirtrons, are known to be derived from introns via a different pathway which bypasses Pasha and Drosha. Some microRNAs are also known to be derived from small nucleolar RNA.

MicroRNA 187 is a protein that in humans is encoded by the MIR187 gene.
MicroRNA 95 is a small non-coding RNA that in humans is encoded by the MIR95 gene.

MicroRNA 34a (miR-34a) is a MicroRNA that in humans is encoded by the MIR34A gene.
DCL1 is a gene in plants that codes for the DCL1 protein, a ribonuclease III enzyme involved in processing microRNA (miRNA). Although DCL1 is named for its homology with the metazoan protein Dicer, its role in miRNA biogenesis is somewhat different, due to substantial differences in miRNA maturation processes between plants and animals. Unlike Dicer, DCL1 is localized to the cell nucleus, where it initiates miRNA processing as the catalytic component of the so-called Dicing complex, which also contains HYL1, a double-stranded RNA binding protein, and a zinc-finger protein known as SE or SERRATE. Within the nucleus, Dicing complexes co-localize in Dicing bodies or D-bodies. In plants, DCL1 is responsible both for processing a primary miRNA to a pre-miRNA, and for then processing the pre-miRNA to a mature miRNA. In animals, the equivalents of these two steps are carried out by different proteins; pri-miRNA processing takes place in the nucleus by the ribonuclease Drosha as part of the Microprocessor complex, and processing to a mature miRNA takes place in the cytoplasm by Dicer to yield a mature miRNA.

MicroRNA 203a is a protein that in humans is encoded by the MIR203A gene.

MicroRNA 148a is a miRNA that in humans is encoded by the MIR148A gene.

MicroRNA 494 is a microRNA sequence which is encoded in humans by the MIR494 gene.

MicroRNA let-7f-2 is a protein that in humans is encoded by the MIRLET7F2 gene.
MicroRNA 517c is a protein that in humans is encoded by the MIR517C gene.















