In biochemistry, a nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the DICER1 gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA).

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.

Ranpirnase is a ribonuclease enzyme found in the oocytes of the Northern Leopard Frog. Ranpirnase is a member of the pancreatic ribonuclease protein superfamily and degrades RNA substrates with a sequence preference for uracil and guanine nucleotides. Along with amphinase, another leopard frog ribonuclease, Ranpirnase has been studied as a potential cancer and antiviral treatment due to its unusual mechanism of cytotoxicity tested against transformed cells and antiviral activity.

Angiogenin (ANG) also known as ribonuclease 5 is a small 123 amino acid protein that in humans is encoded by the ANG gene. Angiogenin is a potent stimulator of new blood vessels through the process of angiogenesis. Ang hydrolyzes cellular RNA, resulting in modulated levels of protein synthesis and interacts with DNA causing a promoter-like increase in the expression of rRNA. Ang is associated with cancer and neurological disease through angiogenesis and through activating gene expression that suppresses apoptosis.
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.
Ribonuclease III (RNase III or RNase C)(BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.

Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the DROSHA gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosis and HIV-1 replication.

The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.

In molecular biology, nuclear ribonuclease P is a ubiquitous endoribonuclease, found in archaea, bacteria and eukarya as well as chloroplasts and mitochondria. Its best characterised enzyme activity is the generation of mature 5′-ends of tRNAs by cleaving the 5′-leader elements of precursor-tRNAs. Cellular RNase Ps are ribonucleoproteins. The RNA from bacterial RNase P retains its catalytic activity in the absence of the protein subunit, i.e. it is a ribozyme. Similarly, archaeal RNase P RNA has been shown to be weakly catalytically active in the absence of its respective protein cofactors. Isolated eukaryotic RNase P RNA has not been shown to retain its catalytic function, but is still essential for the catalytic activity of the holoenzyme. Although the archaeal and eukaryotic holoenzymes have a much greater protein content than the bacterial ones, the RNA cores from all three lineages are homologous—the helices corresponding to P1, P2, P3, P4, and P10/11 are common to all cellular RNase P RNAs. Yet there is considerable sequence variation, particularly among the eukaryotic RNAs.

An exoribonuclease is an exonuclease ribonuclease, which are enzymes that degrade RNA by removing terminal nucleotides from either the 5' end or the 3' end of the RNA molecule. Enzymes that remove nucleotides from the 5' end are called 5'-3' exoribonucleases, and enzymes that remove nucleotides from the 3' end are called 3'-5' exoribonucleases.
Pancreatic ribonuclease family is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles.
Exoribonuclease II is an enzyme. This enzyme catalyses the following chemical reaction
RNase PhyM is a type of endoribonuclease which is sequence specific for single stranded RNAs. It cleaves 3'-end of unpaired A and U residues.
RNase R, or Ribonuclease R, is a 3'-->5' exoribonuclease, which belongs to the RNase II superfamily, a group of enzymes that hydrolyze RNA in the 3' - 5' direction. RNase R has been shown to be involved in selective mRNA degradation, particularly of non stop mRNAs in bacteria. RNase R has homologues in many other organisms.
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.
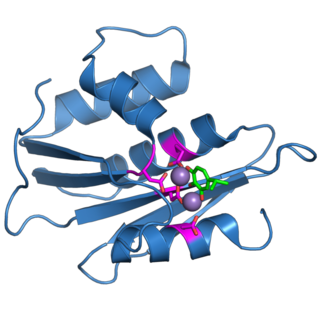
The retroviral ribonuclease H is a catalytic domain of the retroviral reverse transcriptase (RT) enzyme. The RT enzyme is used to generate complementary DNA (cDNA) from the retroviral RNA genome. This process is called reverse transcription. To complete this complex process, the retroviral RT enzymes need to adopt a multifunctional nature. They therefore possess 3 of the following biochemical activities: RNA-dependent DNA polymerase, ribonuclease H, and DNA-dependent DNA polymerase activities. Like all RNase H enzymes, the retroviral RNase H domain cleaves DNA/RNA duplexes and will not degrade DNA or unhybridized RNA.

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.
Ahmed TAE, Udenigwe CC, Gomaa A. Editorial: Biotechnology and Bioengineering Applications for Egg-Derived Biomaterials. Front Bioeng Biotechnol. 2021 Sep 20;9:756058