In chemistry, an acid dissociation constant is a quantitative measure of the strength of an acid in solution. It is the equilibrium constant for a chemical reaction
Cooperative binding occurs in molecular binding systems containing more than one type, or species, of molecule and in which one of the partners is not mono-valent and can bind more than one molecule of the other species. In general, molecular binding is an interaction between molecules that results in a stable physical association between those molecules.
The equilibrium constant of a chemical reaction is the value of its reaction quotient at chemical equilibrium, a state approached by a dynamic chemical system after sufficient time has elapsed at which its composition has no measurable tendency towards further change. For a given set of reaction conditions, the equilibrium constant is independent of the initial analytical concentrations of the reactant and product species in the mixture. Thus, given the initial composition of a system, known equilibrium constant values can be used to determine the composition of the system at equilibrium. However, reaction parameters like temperature, solvent, and ionic strength may all influence the value of the equilibrium constant.

Dissociation in chemistry is a general process in which molecules (or ionic compounds such as salts, or complexes) separate or split into other things such as atoms, ions, or radicals, usually in a reversible manner. For instance, when an acid dissolves in water, a covalent bond between an electronegative atom and a hydrogen atom is broken by heterolytic fission, which gives a proton (H+) and a negative ion. Dissociation is the opposite of association or recombination.
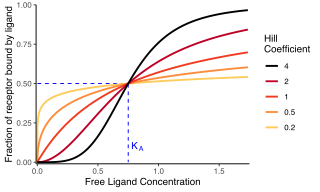
In biochemistry and pharmacology, the Hill equation refers to two closely related equations that reflect the binding of ligands to macromolecules, as a function of the ligand concentration. A ligand is "a substance that forms a complex with a biomolecule to serve a biological purpose", and a macromolecule is a very large molecule, such as a protein, with a complex structure of components. Protein-ligand binding typically changes the structure of the target protein, thereby changing its function in a cell.
In particle physics, the quantum yield of a radiation-induced process is the number of times a specific event occurs per photon absorbed by the system.
Atom transfer radical polymerization (ATRP) is an example of a reversible-deactivation radical polymerization. Like its counterpart, ATRA, or atom transfer radical addition, ATRP is a means of forming a carbon-carbon bond with a transition metal catalyst. Polymerization from this method is called atom transfer radical addition polymerization (ATRAP). As the name implies, the atom transfer step is crucial in the reaction responsible for uniform polymer chain growth. ATRP was independently discovered by Mitsuo Sawamoto and by Krzysztof Matyjaszewski and Jin-Shan Wang in 1995.
The Scatchard equation is an equation used in molecular biology to calculate the affinity and number of binding sites of a receptor for a ligand. It is named after the American chemist George Scatchard.
In biochemistry, avidity refers to the accumulated strength of multiple affinities of individual non-covalent binding interactions, such as between a protein receptor and its ligand, and is commonly referred to as functional affinity. Avidity differs from affinity, which describes the strength of a single interaction. However, because individual binding events increase the likelihood of occurrence of other interactions, avidity should not be thought of as the mere sum of its constituent affinities but as the combined effect of all affinities participating in the biomolecular interaction. A particular important aspect relates to the phenomenon of 'avidity entropy'. Biomolecules often form heterogenous complexes or homogeneous oligomers and multimers or polymers. If clustered proteins form an organized matrix, such as the clathrin-coat, the interaction is described as a matricity.
In biochemistry, receptor–ligand kinetics is a branch of chemical kinetics in which the kinetic species are defined by different non-covalent bindings and/or conformations of the molecules involved, which are denoted as receptor(s) and ligand(s). Receptor–ligand binding kinetics also involves the on- and off-rates of binding.
Equilibrium constants are determined in order to quantify chemical equilibria. When an equilibrium constant K is expressed as a concentration quotient,
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding.
In coordination chemistry, a stability constant is an equilibrium constant for the formation of a complex in solution. It is a measure of the strength of the interaction between the reagents that come together to form the complex. There are two main kinds of complex: compounds formed by the interaction of a metal ion with a ligand and supramolecular complexes, such as host–guest complexes and complexes of anions. The stability constant(s) provide(s) the information required to calculate the concentration(s) of the complex(es) in solution. There are many areas of application in chemistry, biology and medicine.
In chemistry, dissociative substitution describes a reaction pathway by which compounds interchange ligands. The term is typically applied to coordination and organometallic complexes, but resembles the SN1 mechanism in organic chemistry. This pathway can be well described by the cis effect, or the labilization of CO ligands in the cis position. The opposite pathway is associative substitution, being analogous to SN2 pathway. Pathways that are intermediate between the pure dissociative and pure associative pathways are called interchange mechanisms.
In chemistry, binding selectivity is defined with respect to the binding of ligands to a substrate forming a complex. Binding selectivity describes how a ligand may bind more preferentially to one receptor than another. A selectivity coefficient is the equilibrium constant for the reaction of displacement by one ligand of another ligand in a complex with the substrate. Binding selectivity is of major importance in biochemistry and in chemical separation processes.
Equilibrium chemistry is concerned with systems in chemical equilibrium. The unifying principle is that the free energy of a system at equilibrium is the minimum possible, so that the slope of the free energy with respect to the reaction coordinate is zero. This principle, applied to mixtures at equilibrium provides a definition of an equilibrium constant. Applications include acid–base, host–guest, metal–complex, solubility, partition, chromatography and redox equilibria.
Antigen-antibody interaction, or antigen-antibody reaction, is a specific chemical interaction between antibodies produced by B cells of the white blood cells and antigens during immune reaction. The antigens and antibodies combine by a process called agglutination. It is the fundamental reaction in the body by which the body is protected from complex foreign molecules, such as pathogens and their chemical toxins. In the blood, the antigens are specifically and with high affinity bound by antibodies to form an antigen-antibody complex. The immune complex is then transported to cellular systems where it can be destroyed or deactivated.
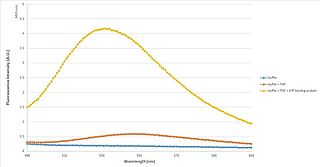
TNP-ATP is a fluorescent molecule that is able to determine whether a protein binds to ATP, and the constants associated with that binding. It is primarily used in fluorescence spectroscopy, but is also very useful as an acceptor molecule in FRET, and as a fluorescent probe in fluorescence microscopy and X-ray crystallography.
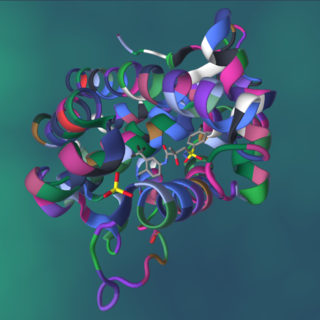
A protein–ligand complex is a complex of a protein bound with a ligand that is formed following molecular recognition between proteins that interact with each other or with other molecules. Formation of a protein-ligand complex is based on molecular recognition between biological macromolecules and ligands, where ligand means any molecule that binds the protein with high affinity and specificity. Molecular recognition is not a process by itself since it is part of a functionally important mechanism involving the essential elements of life like in self-replication, metabolism, and information processing. For example DNA-replication depends on recognition and binding of DNA double helix by helicase, DNA single strand by DNA-polymerase and DNA segments by ligase. Molecular recognition depends on affinity and specificity. Specificity means that proteins distinguish the highly specific binding partner from less specific partners and affinity allows the specific partner with high affinity to remain bound even if there are high concentrations of less specific partners with lower affinity.
Competitive inhibition is interruption of a chemical pathway owing to one chemical substance inhibiting the effect of another by competing with it for binding or bonding. Any metabolic or chemical messenger system can potentially be affected by this principle, but several classes of competitive inhibition are especially important in biochemistry and medicine, including the competitive form of enzyme inhibition, the competitive form of receptor antagonism, the competitive form of antimetabolite activity, and the competitive form of poisoning.













































