Related Research Articles
Asparagine is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group, an α-carboxylic acid group, and a side chain carboxamide, classifying it as a polar, aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it. It is encoded by the codons AAU and AAC.

Glycoproteins are proteins which contain oligosaccharide chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosylation. Secreted extracellular proteins are often glycosylated.
Glycosylation is the reaction in which a carbohydrate, i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule in order to form a glycoconjugate. In biology, glycosylation usually refers to an enzyme-catalysed reaction, whereas glycation may refer to a non-enzymatic reaction.

Haemophilus influenzae is a Gram-negative, non-motile, coccobacillary, facultatively anaerobic, capnophilic pathogenic bacterium of the family Pasteurellaceae. The bacteria are mesophilic and grow best at temperatures between 35 and 37 °C.

The Pasteurellaceae comprise a large family of Gram-negative bacteria. Most members live as commensals on mucosal surfaces of birds and mammals, especially in the upper respiratory tract. Pasteurellaceae are typically rod-shaped, and are a notable group of facultative anaerobes. Their biochemical characteristics can be distinguished from the related Enterobacteriaceae by the presence of oxidase, and from most other similar bacteria by the absence of flagella.

Oligosaccharyltransferase or OST (EC 2.4.1.119) is a membrane protein complex that transfers a 14-sugar oligosaccharide from dolichol to nascent protein. It is a type of glycosyltransferase. The sugar Glc3Man9GlcNAc2 (where Glc=Glucose, Man=Mannose, and GlcNAc=N-acetylglucosamine) is attached to an asparagine (Asn) residue in the sequence Asn-X-Ser or Asn-X-Thr where X is any amino acid except proline. This sequence is called a glycosylation sequon. The reaction catalyzed by OST is the central step in the N-linked glycosylation pathway.

Glycosyltransferases are enzymes that establish natural glycosidic linkages. They catalyze the transfer of saccharide moieties from an activated nucleotide sugar to a nucleophilic glycosyl acceptor molecule, the nucleophile of which can be oxygen- carbon-, nitrogen-, or sulfur-based.

Ribophorins are dome shaped transmembrane glycoproteins which are located in the membrane of the rough endoplasmic reticulum, but are absent in the membrane of the smooth endoplasmic reticulum. There are two types of ribophorines: ribophorin I and II. These act in the protein complex oligosaccharyltransferase (OST) as two different subunits of the named complex. Ribophorin I and II are only present in eukaryote cells.
In enzymology, a dolichyl-diphosphooligosaccharide–protein glycotransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a protein N-acetylglucosaminyltransferase is an enzyme that catalyzes the chemical reaction

Alpha-1,3/1,6-mannosyltransferase ALG2 is an enzyme that is encoded by the ALG2 gene. Mutations in the human gene are associated with congenital defects in glycosylation The protein encoded by the ALG2 gene belongs to two classes of enzymes: GDP-Man:Man1GlcNAc2-PP-dolichol alpha-1,3-mannosyltransferase and GDP-Man:Man2GlcNAc2-PP-dolichol alpha-1,6-mannosyltransferase.

Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 2, also called ribophorin ǁ is an enzyme that in humans is encoded by the RPN2 gene.

Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase is an enzyme that in humans is encoded by the ALG8 gene.

Dolichyl-P-Man:Man(7)GlcNAc(2)-PP-dolichyl-alpha-1,6-mannosyltransferase is an enzyme that in humans is encoded by the ALG12 gene.

Dolichyl-diphosphooligosaccharide—protein glycosyltransferase 48 kDa subunit is an enzyme that in humans is encoded by the DDOST gene.

Chitobiosyldiphosphodolichol beta-mannosyltransferase is an enzyme that is encoded by ALG1 whose structure and function has been conserved from lower to higher organisms.
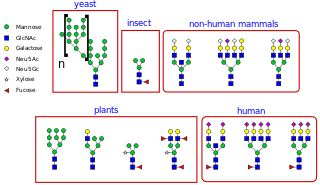
N-linked glycosylation, is the attachment of an oligosaccharide, a carbohydrate consisting of several sugar molecules, sometimes also referred to as glycan, to a nitrogen atom, in a process called N-glycosylation, studied in biochemistry. The resulting protein is called an N-linked glycan, or simply an N-glycan.

In molecular biology, trimeric autotransporter adhesins (TAAs), are proteins found on the outer membrane of Gram-negative bacteria. Bacteria use TAAs in order to infect their host cells via a process called cell adhesion. TAAs also go by another name, oligomeric coiled-coil adhesins, which is shortened to OCAs. In essence, they are virulence factors, factors that make the bacteria harmful and infective to the host organism.

Protein O-GlcNAc transferase also known as OGT or O-linked N-acetylglucosaminyltransferase is an enzyme that in humans is encoded by the OGT gene. OGT catalyzes the addition of the O-GlcNAc post-translational modification to proteins.
The Nicotinamide Ribonucleoside (NR) Uptake Permease (PnuC) Family is a family of transmembrane transporters that is part of the TOG superfamily. Close PnuC homologues are found in a wide range of Gram-negative and Gram-positive bacteria, archaea and eukaryotes.
References
- 1 2 3 Nothaft & Szymanski 2013, p. 6916.
- ↑ Choi et al. 2010, p. 2.
- 1 2 Song et al. 2017, p. 8856.
- 1 2 3 Naegeli & Aebi 2015, p. 11.
- ↑ Nothaft & Szymanski 2013, p. 6912.
- 1 2 3 4 Grass S, Lichti CF, Townsend RR, Gross J, St Geme JW (May 2010). "The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin". PLOS Pathogens. 6 (5): e1000919. doi: 10.1371/journal.ppat.1000919 . PMC 2877744 . PMID 20523900.
- 1 2 3 4 Gawthorne et al. 2014, p. 633.
- 1 2 3 4 5 6 7 8 Naegeli et al. 2014, p. 24522.
- 1 2 Naegeli et al. 2014, p. 24521.
- ↑ Piniello, Macías-León & Miyazaki 2023, p. 8.
- ↑ Bause & Legler 1981, p. 644.
- 1 2 3 4 5 6 7 Schwarz et al. 2011, p. 35273.
- 1 2 3 4 5 6 7 Schwarz et al. 2011, p. 35267.
- 1 2 Piniello, Macías-León & Miyazaki 2023, p. 2.
- ↑ Song et al. 2017, p. 8861.
- ↑ Bause E (February 1983). "Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes". The Biochemical Journal. 209 (2): 331–336. doi:10.1042/bj2090331. PMC 1154098 . PMID 6847620.
- 1 2 Naegeli et al. 2014, p. 24524.
- ↑ Naegeli et al. 2014, p. 24530.
- ↑ Choi et al. 2010, p. 7.
- 1 2 Naegeli et al. 2014, p. 2171.
- ↑ Kawai et al. 2011, p. 38553.
- 1 2 McCann & St Geme 2014, p. 2.
- 1 2 Choi et al. 2010, p. 1.
- 1 2 3 4 Naegeli et al. 2014, p. 2170.
- ↑ Bause & Legler 1981, p. 639.
- 1 2 Naegeli & Aebi 2015, p. 4.
- ↑ Grass et al. 2003, p. 737.
- ↑ Schwarz et al. 2011, p. 35269.
- ↑ Gawthorne et al. 2014, p. 636.
- 1 2 Song et al. 2017, p. 8857.
- 1 2 3 McCann & St Geme 2014, p. 3.
- ↑ Naegeli & Aebi 2015, p. 12.
- ↑ Naegeli et al. 2014, p. 2172.
- ↑ Keys TG, Wetter M, Hang I, Rutschmann C, Russo S, Mally M, et al. (November 2017). "A biosynthetic route for polysialylating proteins in Escherichia coli". Metabolic Engineering. 44: 293–301. doi: 10.1016/j.ymben.2017.10.012 . PMID 29101090.
- ↑ Naegeli et al. 2014, p. 2173.
- 1 2 Naegeli et al. 2014, p. 2178.
- 1 2 Naegeli et al. 2014, p. 24531.
- ↑ Gawthorne et al. 2014, p. 634.
- ↑ Kawai et al. 2011, p. 38547.
- ↑ Kawai et al. 2011, p. 38549,38550.
- ↑ Cuccui et al. 2017, p. 2.
- ↑ Cuccui et al. 2017, p. 10.
- 1 2 3 Rempe et al. 2015, p. 5.
- 1 2 Rempe et al. 2015, p. 4.
- 1 2 3 McCann & St Geme 2014, p. 1.
- ↑ Rempe et al. 2015, p. 2.
- ↑ Gawthorne et al. 2014, p. 637,638.
- ↑ Grass et al. 2003, p. 742.
- ↑ Kawai et al. 2011, p. 38546.
- 1 2 Valguarnera E, Kinsella RL, Feldman MF (August 2016). "Sugar and Spice Make Bacteria Not Nice: Protein Glycosylation and Its Influence in Pathogenesis". Journal of Molecular Biology. 428 (16): 3206–3220. doi:10.1016/j.jmb.2016.04.013. PMID 27107636.
- ↑ Rempe et al. 2015, p. 3.
- ↑ Rempe et al. 2015, p. 6.
Sources
- Bause E, Legler G (June 1981). "The role of the hydroxy amino acid in the triplet sequence Asn-Xaa-Thr(Ser) for the N-glycosylation step during glycoprotein biosynthesis". The Biochemical Journal. 195 (3): 639–644. doi:10.1042/bj1950639. PMC 1162935 . PMID 7316978.
- Choi KJ, Grass S, Paek S, St Geme JW, Yeo HJ (December 2010). "The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin". PLOS ONE. 5 (12): e15888. Bibcode:2010PLoSO...515888C. doi: 10.1371/journal.pone.0015888 . PMC 3012730 . PMID 21209858.
- Cuccui J, Terra VS, Bossé JT, Naegeli A, Abouelhadid S, Li Y, et al. (January 2017). "The N-linking glycosylation system from Actinobacillus pleuropneumoniae is required for adhesion and has potential use in glycoengineering". Open Biology. 7 (1): 160212. doi:10.1098/rsob.160212. PMC 5303269 . PMID 28077594.
- Gawthorne JA, Tan NY, Bailey UM, Davis MR, Wong LW, Naidu R, et al. (March 2014). "Selection against glycosylation sites in potential target proteins of the general HMWC N-glycosyltransferase in Haemophilus influenzae". Biochemical and Biophysical Research Communications. 445 (3): 633–638. doi:10.1016/j.bbrc.2014.02.044. PMID 24565833.
- Grass S, Buscher AZ, Swords WE, Apicella MA, Barenkamp SJ, Ozchlewski N, St Geme JW (May 2003). "The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis". Molecular Microbiology. 48 (3): 737–751. doi:10.1046/j.1365-2958.2003.03450.x. PMID 12694618. S2CID 25667209.
- Kawai F, Grass S, Kim Y, Choi KJ, St Geme JW, Yeo HJ (November 2011). "Structural insights into the glycosyltransferase activity of the Actinobacillus pleuropneumoniae HMW1C-like protein". The Journal of Biological Chemistry. 286 (44): 38546–38557. doi: 10.1074/jbc.M111.237602 . PMC 3207471 . PMID 21908603.
- McCann JR, St Geme JW (April 2014). "The HMW1C-like glycosyltransferases--an enzyme family with a sweet tooth for simple sugars". PLOS Pathogens. 10 (4): e1003977. doi: 10.1371/journal.ppat.1003977 . PMC 3983070 . PMID 24722584.
- Naegeli A, Aebi M (2015). "Current Approaches to Engineering N-Linked Protein Glycosylation in Bacteria". Glyco-Engineering. Methods in Molecular Biology. Vol. 1321. Humana Press, New York, NY. pp. 3–16. doi:10.1007/978-1-4939-2760-9_1. ISBN 978-1-4939-2759-3. PMID 26082211.
- Naegeli A, Michaud G, Schubert M, Lin CW, Lizak C, Darbre T, et al. (August 2014). "Substrate specificity of cytoplasmic N-glycosyltransferase". The Journal of Biological Chemistry. 289 (35): 24521–24532. doi: 10.1074/jbc.M114.579326 . PMC 4148877 . PMID 24962585.
- Naegeli A, Neupert C, Fan YY, Lin CW, Poljak K, Papini AM, et al. (January 2014). "Molecular analysis of an alternative N-glycosylation machinery by functional transfer from Actinobacillus pleuropneumoniae to Escherichia coli". The Journal of Biological Chemistry. 289 (4): 2170–2179. doi: 10.1074/jbc.M113.524462 . PMC 3900963 . PMID 24275653.
- Nothaft H, Szymanski CM (March 2013). "Bacterial protein N-glycosylation: new perspectives and applications". The Journal of Biological Chemistry. 288 (10): 6912–6920. doi: 10.1074/jbc.R112.417857 . PMC 3591601 . PMID 23329827.
- Piniello B, Macías-León J, Miyazaki S (2023). "Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase". Nat Commun. 14. doi:10.1038/s41467-023-41238-1.
- Rempe KA, Spruce LA, Porsch EA, Seeholzer SH, Nørskov-Lauritsen N, St Geme JW (August 2015). "Unconventional N-Linked Glycosylation Promotes Trimeric Autotransporter Function in Kingella kingae and Aggregatibacter aphrophilus". mBio. 6 (4): e01206–15. doi:10.1128/mBio.01206-15. PMC 4550697 . PMID 26307167.
- Schwarz F, Fan YY, Schubert M, Aebi M (October 2011). "Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence". The Journal of Biological Chemistry. 286 (40): 35267–35274. doi: 10.1074/jbc.M111.277160 . PMC 3186387 . PMID 21852240.
- Song Q, Wu Z, Fan Y, Song W, Zhang P, Wang L, et al. (May 2017). "Production of homogeneous glycoprotein with multisite modifications by an engineered N-glycosyltransferase mutant". The Journal of Biological Chemistry. 292 (21): 8856–8863. doi: 10.1074/jbc.M117.777383 . PMC 5448120 . PMID 28381551.