
Bacillus is a genus of Gram-positive, rod-shaped bacteria, a member of the phylum Bacillota, with 266 named species. The term is also used to describe the shape (rod) of other so-shaped bacteria; and the plural Bacilli is the name of the class of bacteria to which this genus belongs. Bacillus species can be either obligate aerobes which are dependent on oxygen, or facultative anaerobes which can survive in the absence of oxygen. Cultured Bacillus species test positive for the enzyme catalase if oxygen has been used or is present.

A microbiological culture, or microbial culture, is a method of multiplying microbial organisms by letting them reproduce in predetermined culture medium under controlled laboratory conditions. Microbial cultures are foundational and basic diagnostic methods used as research tools in molecular biology.

Polymyxins are antibiotics. Polymyxins B and E are used in the treatment of Gram-negative bacterial infections. They work mostly by breaking up the bacterial cell membrane. They are part of a broader class of molecules called nonribosomal peptides.
Collaborative intelligence characterizes multi-agent, distributed systems where each agent, human or machine, is autonomously contributing to a problem solving network. Collaborative autonomy of organisms in their ecosystems makes evolution possible. Natural ecosystems, where each organism's unique signature is derived from its genetics, circumstances, behavior and position in its ecosystem, offer principles for design of next generation social networks to support collaborative intelligence, crowdsourcing individual expertise, preferences, and unique contributions in a problem solving process.

An endophyte is an endosymbiont, often a bacterium or fungus, that lives within a plant for at least part of its life cycle without causing apparent disease. Endophytes are ubiquitous and have been found in all species of plants studied to date; however, most of the endophyte/plant relationships are not well understood. Some endophytes may enhance host growth, nutrient acquisition and improve the plant's ability to tolerate abiotic stresses, such as drought and decrease biotic stresses by enhancing plant resistance to insects, pathogens and herbivores. Although endophytic bacteria and fungi are frequently studied, endophytic archaea are increasingly being considered for their role in plant growth promotion as part of the core microbiome of a plant.
Diazotrophs are bacteria and archaea that fix gaseous nitrogen in the atmosphere into a more usable form such as ammonia.
Microbial intelligence is the intelligence shown by microorganisms. The concept encompasses complex adaptive behavior shown by single cells, and altruistic or cooperative behavior in populations of like or unlike cells mediated by chemical signalling that induces physiological or behavioral changes in cells and influences colony structures.
Paenibacillus polymyxa, also known as Bacillus polymyxa, is a Gram-positive bacterium capable of fixing nitrogen. It is found in soil, plant tissues, marine sediments and hot springs. It may have a role in forest ecosystems and potential future applications as a biofertilizer and biocontrol agent in agriculture.

Bacteria are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
Pseudomonas lini is a fluorescent, Gram-negative, rod-shaped bacterium isolated from rhizospheric soil in France. The type strain is CFBP 5737, though there are also eight other strains known. This bacterium has also been isolated from endophytic tissues of lodgepole pine trees growing on gravel mining sites with potential to perform biological nitrogen fixation and plant growth promotion.
Pseudomonas migulae is a fluorescent, Gram-negative, rod-shaped bacterium isolated from natural mineral waters in France. This bacterium has also been isolated from endophytic tissues of lodgepole pine trees growing on gravel mining sites with potential to perform biological nitrogen fixation and plant growth promotion. Based on 16S rRNA analysis, P. migulae has been placed in the P. fluorescens group.
Paraburkholderia phytofirmans is a species of bacteria. They have been reported to colonize endophytic tissues of hybrid spruce and lodgepole pine with a strong potential to perform biological nitrogen fixation and plant growth promotion.
Caballeronia sordidicola is a species of bacteria which has been reported to perform biological nitrogen fixation and promote plant growth

A prokaryote is a single-cell organism whose cell lacks a nucleus and other membrane-bound organelles. The word prokaryote comes from the Ancient Greek πρό 'before' and κάρυον 'nut, kernel'. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: Bacteria and Archaea. Organisms with nuclei are placed in a third domain, Eukaryota.

Paenibacillus vortex is a species of pattern-forming bacteria, first discovered in the early 1990s by Eshel Ben-Jacob's group at Tel Aviv University. It is a social microorganism that forms colonies with complex and dynamic architectures. P. vortex is mainly found in heterogeneous and complex environments, such as the rhizosphere, the soil region directly influenced by plant roots.

Paenibacillus dendritiformis is a species of pattern-forming bacteria, first discovered in the early 90s by Eshel Ben-Jacob's group. It is a social microorganism that forms colonies with complex and dynamic architectures. The genus Paenibacillus comprises facultative anaerobic, endospore-forming bacteria originally included within the genus Bacillus and then reclassified as a separate genus in 1993. Bacteria belonging to this genus have been detected in a variety of environments such as: soil, water, rhizosphere, vegetable matter, forage and insect larvae.
Caballeronia udeis is a bacterium from the genus Caballeronia and family Burkholderiaceae which has been reported to perform biological nitrogen fixation and promote plant growth
Paraburkholderia is a genus of Pseudomonadota that are gram negative, slightly curved rods that are motile by means of flagella. They have been reported to colonize endophytic tissues of hybrid spruce and lodgepole pine with a strong potential to perform biological nitrogen fixation and plant growth promotion. Unlike Burkholderia species, Paraburkholderia members are not commonly associated with human infection. Paraburkholderia members form a monophyletic clade within the Burkholderiaceae family, which is what prompted their distinction as a genus independent from Burkholderia species, in combination with the finding of robust conserved signature indels which are unique to Paraburkholderia species, and are lacking in members of the genus Burkholderia. These CSIs distinguish the genus from all other bacteria. Additionally, the CSIs that were found to be shared by Burkholderia species are absent in Paraburkholderia, providing evidence of separate lineages.
Caballeronia is a genus of bacteria from the family of Burkholderiaceae which has been reported to perform biological nitrogen fixation and promote plant growth
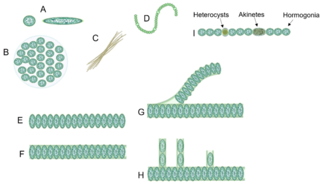
Cyanobacterial morphology refers to the form or shape of cyanobacteria. Cyanobacteria are a large and diverse phylum of bacteria defined by their unique combination of pigments and their ability to perform oxygenic photosynthesis.











