The cell cycle, or cell-division cycle, is the series of events that take place in a cell that causes it to divide into two daughter cells. These events include the duplication of its DNA and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division.

Mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis is an equational division which gives rise to genetically identical cells in which the total number of chromosomes is maintained. Mitosis is preceded by the S phase of interphase and is followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic phase of a cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.

The G1 phase, gap 1 phase, or growth 1 phase, is the first of four phases of the cell cycle that takes place in eukaryotic cell division. In this part of interphase, the cell synthesizes mRNA and proteins in preparation for subsequent steps leading to mitosis. G1 phase ends when the cell moves into the S phase of interphase. Around 30 to 40 percent of cell cycle time is spent in the G1 phase.
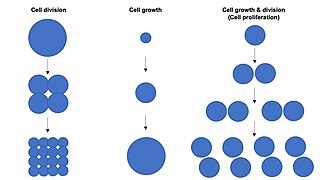
Cell growth refers to an increase in the total mass of a cell, including both cytoplasmic, nuclear and organelle volume. Cell growth occurs when the overall rate of cellular biosynthesis is greater than the overall rate of cellular degradation.

Thymidine kinase is an enzyme, a phosphotransferase : 2'-deoxythymidine kinase, ATP-thymidine 5'-phosphotransferase, EC 2.7.1.21. It can be found in most living cells. It is present in two forms in mammalian cells, TK1 and TK2. Certain viruses also have genetic information for expression of viral thymidine kinases. Thymidine kinase catalyzes the reaction:
A mitogen is a small bioactive protein or peptide that induces a cell to begin cell division, or enhances the rate of division (mitosis). Mitogenesis is the induction (triggering) of mitosis, typically via a mitogen. The mechanism of action of a mitogen is that it triggers signal transduction pathways involving mitogen-activated protein kinase (MAPK), leading to mitosis.
Mitotic index is defined as the ratio between the number of a population's cells undergoing mitosis to its total number of cells.
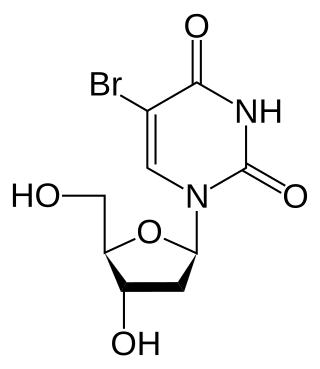
Bromodeoxyuridine is a synthetic nucleoside analogue with a chemical structure similar to thymidine. BrdU is commonly used to study cell proliferation in living tissues and has been studied as a radiosensitizer and diagnostic tool in people with cancer.

Cyclin B is a member of the cyclin family. Cyclin B is a mitotic cyclin. The amount of cyclin B and the activity of the cyclin B-Cdk complex rise through the cell cycle until mitosis, where they fall abruptly due to degradation of cyclin B. The complex of Cdk and cyclin B is called maturation promoting factor or mitosis promoting factor (MPF).
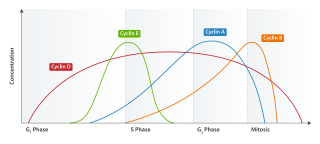
Cyclin E is a member of the cyclin family.

Aurora kinase A also known as serine/threonine-protein kinase 6 is an enzyme that in humans is encoded by the AURKA gene.

Antigen Kiel 67, also known as Ki-67 or MKI67, is a protein that in humans is encoded by the MKI67 gene.
Cell synchronization is a process by which cells in a culture at different stages of the cell cycle are brought to the same phase. Cell synchrony is a vital process in the study of cells progressing through the cell cycle as it allows population-wide data to be collected rather than relying solely on single-cell experiments. The types of synchronization are broadly categorized into two groups; physical fractionization and chemical blockade.

Cyclin-dependent kinase inhibitor 1B (p27Kip1) is an enzyme inhibitor that in humans is encoded by the CDKN1B gene. It encodes a protein which belongs to the Cip/Kip family of cyclin dependent kinase (Cdk) inhibitor proteins. The encoded protein binds to and prevents the activation of cyclin E-CDK2 or cyclin D-CDK4 complexes, and thus controls the cell cycle progression at G1. It is often referred to as a cell cycle inhibitor protein because its major function is to stop or slow down the cell division cycle.
G2/mitotic-specific cyclin-B1 is a protein that in humans is encoded by the CCNB1 gene.
Thymidine kinase 1, soluble, is a human thymidine kinase.
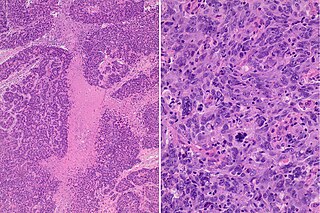
The basal-like carcinoma is a recently proposed subtype of breast cancer defined by its gene expression and protein expression profile.

SCL-interrupting locus protein is a protein that in humans is encoded by the STIL gene. STIL is present in many different cell types and is essential for centriole biogenesis. This gene encodes a cytoplasmic protein implicated in regulation of the mitotic spindle checkpoint, a regulatory pathway that monitors chromosome segregation during cell division to ensure the proper distribution of chromosomes to daughter cells. The protein is phosphorylated in mitosis and in response to activation of the spindle checkpoint, and disappears when cells transition to G1 phase. It interacts with a mitotic regulator, and its expression is required to efficiently activate the spindle checkpoint.

Mitotic catastrophe has been defined as either a cellular mechanism to prevent potentially cancerous cells from proliferating or as a mode of cellular death that occurs following improper cell cycle progression or entrance. Mitotic catastrophe can be induced by prolonged activation of the spindle assembly checkpoint, errors in mitosis, or DNA damage and operates to prevent genomic instability. It is a mechanism that is being researched as a potential therapeutic target in cancers, and numerous approved therapeutics induce mitotic catastrophe.
Thymidine kinase is an enzyme, a phosphotransferase : 2'-deoxythymidine kinase, ATP-thymidine 5'-phosphotransferase, EC 2.7.1.21 that catalyzes the reaction: