Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950 when she noticed that streaks of mixtures of two E. coli strains, one of which treated with ultraviolet light, was "nibbled and plaqued". The wild type of this virus has a temperate lifecycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase ; mutant strains are unable to lysogenize cells – instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

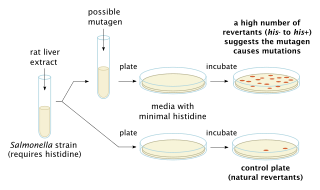
In genetics, a mutagen is a physical or chemical agent that changes the genetic material, usually DNA, of an organism and thus increases the frequency of mutations above the natural background level. As many mutations can cause cancer, mutagens are therefore also likely to be carcinogens, although not always necessarily so. All mutagens have characteristic mutational signatures with some chemicals becoming mutagenic through cellular processes. Not all mutations are caused by mutagens: so-called "spontaneous mutations" occur due to spontaneous hydrolysis, errors in DNA replication, repair and recombination.

β-galactosidase, also called lactase, beta-gal or β-gal, is a glycoside hydrolase enzyme that catalyzes the hydrolysis of β-galactosides into monosaccharides through the breaking of a glycosidic bond. β-galactosides include carbohydrates containing galactose where the glycosidic bond lies above the galactose molecule. Substrates of different β-galactosidases include ganglioside GM1, lactosylceramides, lactose, and various glycoproteins.

François Jacob was a French biologist who, together with Jacques Monod, originated the idea that control of enzyme levels in all cells occurs through regulation of transcription. He shared the 1965 Nobel Prize in Medicine with Jacques Monod and André Lwoff.

The lac operon is an operon required for the transport and metabolism of lactose in Escherichia coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.
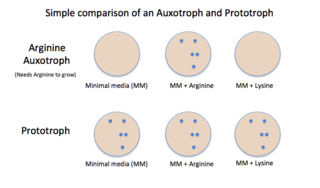
Auxotrophy is the inability of an organism to synthesize a particular organic compound required for its growth. An auxotroph is an organism that displays this characteristic; auxotrophic is the corresponding adjective. Auxotrophy is the opposite of prototrophy, which is characterized by the ability to synthesize all the compounds needed for growth.

Allolactose is a disaccharide similar to lactose. It consists of the monosaccharides D-galactose and D-glucose linked through a β1-6 glycosidic linkage instead of the β1-4 linkage of lactose. It may arise from the occasional transglycosylation of lactose by β-galactosidase.

Isopropyl β-D-1-thiogalactopyranoside (IPTG) is a molecular biology reagent. This compound is a molecular mimic of allolactose, a lactose metabolite that triggers transcription of the lac operon, and it is therefore used to induce protein expression where the gene is under the control of the lac operator.
In molecular biology, an inducer is a molecule that regulates gene expression. An inducer can bind to protein repressors or activators.

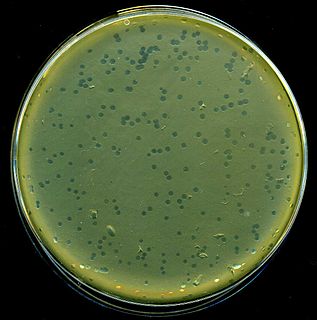
X-gal is an organic compound consisting of galactose linked to a substituted indole. The compound was synthesized by Jerome Horwitz and collaborators in Detroit, MI, in 1964. The formal chemical name is often shortened to less accurate but also less cumbersome phrases such as bromochloroindoxyl galactoside. The X from indoxyl may be the source of the X in the X-gal contraction. X-gal is often used in molecular biology to test for the presence of an enzyme, β-galactosidase. It is also used to detect activity of this enzyme in histochemistry and bacteriology. X-gal is one of many indoxyl glycosides and esters that yield insoluble blue compounds similar to indigo as a result of enzyme-catalyzed hydrolysis.
Diauxie is a Greek word coined by Jacques Monod to mean two growth phases. The word is used in English in cell biology to describe the growth phases of a microorganism in batch culture as it metabolizes a mixture of two sugars. Rather than metabolizing the two available sugars simultaneously, microbial cells commonly consume them in a sequential pattern, resulting in two separate growth phases.
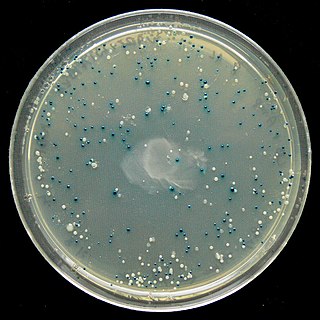
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. DNA of interest is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used.
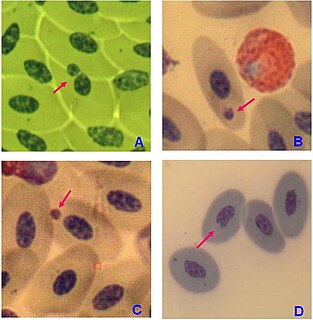
A micronucleus test is a test used in toxicological screening for potential genotoxic compounds. The assay is now recognized as one of the most successful and reliable assays for genotoxic carcinogens, i.e., carcinogens that act by causing genetic damage and is the OECD guideline for the testing of chemicals. There are two major versions of this test, one in vivo and the other in vitro.
Carbon catabolite repression, or simply catabolite repression, is an important part of global control system of various bacteria and other micro-organisms. Catabolite repression allows micro-organisms to adapt quickly to a preferred carbon and energy source first. This is usually achieved through inhibition of synthesis of enzymes involved in catabolism of carbon sources other than the preferred one. The catabolite repression was first shown to be initiated by glucose and therefore sometimes referred to as the glucose effect. However, the term "glucose effect" is actually a misnomer since other carbon sources are known to induce catabolite repression.
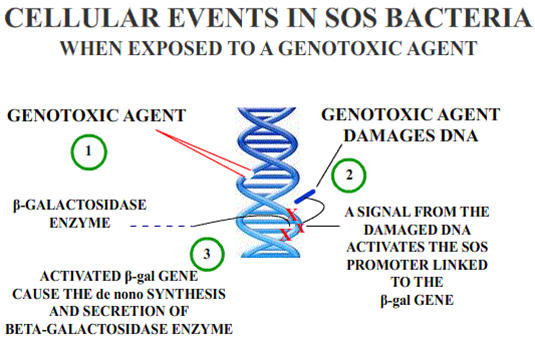
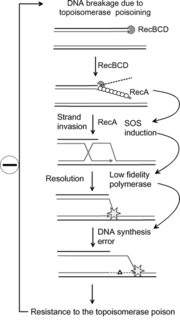
The Umu Chromotest, first developed and published by Oda et al., is a biological assay (bioassay) to assess the genotoxic potential of chemical compounds. It is based on the ability of DNA-damaging agents to induce the expression of the umu operon. In connection with the damage inducible (din) genes recA, lexA and umuD, the umuC gene is essentially involved in bacterial mutagenesis through the SOS response.
E-SCREEN is a cell proliferation assay based on the enhanced proliferation of human breast cancer cells (MCF-7) in the presence of estrogen active substances. The E-SCREEN test is a reliable tool to easily and rapidly assess estrogenic activity of suspected xenoestrogens. This bioassay that measures estrogen-induced increase of the number of human breast cancer cell, which is biologically equivalent to the increase of mitotic activity in tissues of the genital tract, was originally developed by Soto et al.