Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Molybdopterins are a class of cofactors found in most molybdenum-containing and all tungsten-containing enzymes. Synonyms for molybdopterin are: MPT and pyranopterin-dithiolate. The nomenclature for this biomolecule can be confusing: Molybdopterin itself contains no molybdenum; rather, this is the name of the ligand that will bind the active metal. After molybdopterin is eventually complexed with molybdenum, the complete ligand is usually called molybdenum cofactor.

Sulfite oxidase is an enzyme in the mitochondria of all eukaryotes, with exception of the yeasts. It oxidizes sulfite to sulfate and, via cytochrome c, transfers the electrons produced to the electron transport chain, allowing generation of ATP in oxidative phosphorylation. This is the last step in the metabolism of sulfur-containing compounds and the sulfate is excreted.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.

Nitrate reductases are molybdoenzymes that reduce nitrate to nitrite. This reaction is critical for the production of protein in most crop plants, as nitrate is the predominant source of nitrogen in fertilized soils.
Nitrate reductase (NADH) (EC 1.7.1.1, assimilatory nitrate reductase, NADH-nitrate reductase, NADH-dependent nitrate reductase, assimilatory NADH: nitrate reductase, nitrate reductase (NADH2), NADH2:nitrate oxidoreductase) is an enzyme with systematic name nitrite:NAD+ oxidoreductase. This enzyme catalyzes the following chemical reaction
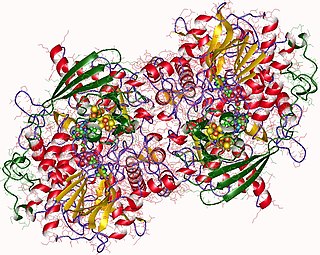
Adenylyl-sulfate reductase is an enzyme that catalyzes the chemical reaction of the reduction of adenylyl-sulfate/adenosine-5'-phosphosulfate (APS) to sulfite through the use of an electron donor cofactor. The products of the reaction are AMP and sulfite, as well as an oxidized electron donor cofactor.
Adenylyl-sulfate reductase (glutathione) is an enzyme that catalyzes the chemical reaction
In enzymology, a ferredoxin—nitrite reductase (EC 1.7.7.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydrogensulfite reductase (EC 1.8.99.3) is an enzyme that catalyzes the chemical reaction
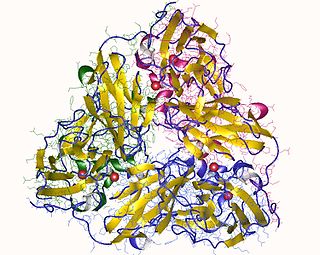
In enzymology, a nitrite reductase (NO-forming) (EC 1.7.2.1) is an enzyme that catalyzes the chemical reaction

Sulfite reductases (EC 1.8.99.1) are enzymes that participate in sulfur metabolism. They catalyze the reduction of sulfite to hydrogen sulfide and water. Electrons for the reaction are provided by a dissociable molecule of either NADPH, bound flavins, or ferredoxins.

Thiosulfate dehydrogenase is an enzyme that catalyzes the chemical reaction:
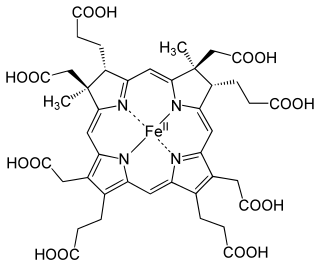
The enzyme sirohydrochlorin ferrochelatase (EC 4.99.1.4) catalyzes the following reaction:
Sulfur is metabolized by all organisms, from bacteria and archaea to plants and animals. Sulfur can have an oxidation state from -2 to +6 and is reduced or oxidized by a diverse range of organisms. The element is present in proteins, sulfate esters of polysaccharides, steroids, phenols, and sulfur-containing coenzymes.

F430 is the cofactor (sometimes called the coenzyme) of the enzyme methyl coenzyme M reductase (MCR). MCR catalyzes the reaction EC 2.8.4.1 that releases methane in the final step of methanogenesis:

Dissimilatory sulfate reduction is a form of anaerobic respiration that uses sulfate as the terminal electron acceptor to produce hydrogen sulfide. This metabolism is found in some types of bacteria and archaea which are often termed sulfate-reducing organisms. The term "dissimilatory" is used when hydrogen sulfide is produced in an anaerobic respiration process. By contrast, the term "assimilatory" would be used in relation to the biosynthesis of organosulfur compounds, even though hydrogen sulfide may be an intermediate.
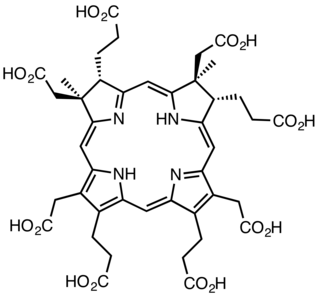
Sirohydrochlorin is a tetrapyrrole macrocyclic metabolic intermediate in the biosynthesis of sirohaem, the iron-containing prosthetic group in sulfite reductase enzymes. It is also the biosynthetic precursor to cofactor F430, an enzyme which catalyzes the release of methane in the final step of methanogenesis.
Dissimilatory sulfite reductase is an enzyme that participates in sulfur metabolism in dissimilatory sulfate reduction.












