
Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. They are characterized by their cell envelopes, which are composed of a thin peptidoglycan cell wall sandwiched between an inner membrane (cytoplasmic), and an outer membrane.
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer (sacculus) that surrounds the bacterial cytoplasmic membrane. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.

The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria, between cell wall and the plasma membrane.
The cell envelope comprises the inner cell membrane and the cell wall of a bacterium. In gram-negative bacteria an outer membrane is also included. This envelope is not present in the Mollicutes where the cell wall is absent.
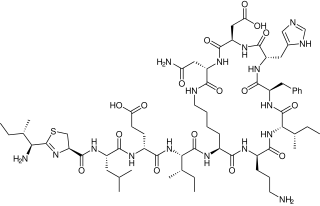
Bacitracin is a polypeptide antibiotic. It is a mixture of related cyclic peptides produced by Bacillus licheniformis bacteria, that was first isolated from the variety "Tracy I" in 1945. These peptides disrupt gram-positive bacteria by interfering with cell wall and peptidoglycan synthesis.

The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.
The bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.
In enzymology, an undecaprenyl-diphosphatase (EC 3.6.1.27) is an enzyme that catalyzes the chemical reaction
Peptidoglycan glycosyltransferase is an enzyme used in the biosynthesis of peptidoglycan. It transfers a disaccharide-peptide from a donor substrate to synthesize a glycan chain.
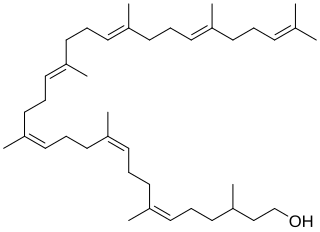
Bactoprenol also known as dolichol-11 and C55-isoprenyl alcohol (C55-OH) is a lipid first identified in certain species of lactobacilli. It is a hydrophobic alcohol that plays a key role in the growth of cell walls (peptidoglycan) in Gram-positive bacteria.
C55-isoprenyl pyrophosphate is an essential molecule involved in construction of the bacterial peptidoglycan cell wall. It is a receptor found in the plasma membrane of bacteria that allows glycan tetrapeptide monomers synthesized in the cell cytoplasm to translocate to the periplasmic space.
UDP-N-acetylglucosamine—undecaprenyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:ditrans,octacis-undecaprenyl phosphate N-acetyl-alpha-D-glucosaminephosphotransferase. This enzyme catalyses the following chemical reaction
Eleftheria terrae is a recently discovered Gram-negative bacterium. E. terrae is a temporary name for the organism, as it was only discovered in 2014 and is still undergoing scientific study. It was found to produce a previously unknown antibiotic named teixobactin. The discovery of E. terrae could represent a new age of antibiotics, as teixobactin is the first new antibiotic discovered since the synthetic era of the 1980s. Prior research has indicated that other uncultivable bacteria like E. terrae have potential in the development of new antimicrobial agents.

Lipid II is a precursor molecule in the synthesis of the cell wall of bacteria. It is a peptidoglycan, which is amphipathic and named for its bactoprenol hydrocarbon chain, which acts as a lipid anchor, embedding itself in the bacterial cell membrane. Lipid II must translocate across the cell membrane to deliver and incorporate its disaccharide-pentapeptide "building block" into the peptidoglycan mesh. Lipid II is the target of several antibiotics.
The bacterial murein precursor exporter (MPE) family is a member of the cation diffusion facilitator (CDF) superfamily of membrane transporters. Members of the MPE family are found in a large variety of Gram-negative and Gram-positive bacteria and facilitate the translocation of lipid-linked murein precursors. A representative list of proteins belonging to the MPE family can be found in the Transporter Classification Database.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family

First described in 1965, the moenomycins are a family of phosphoglycolipid antibiotics, metabolites of the bacterial genus Streptomyces. Moenomycin A is the founding member of the antibiotic family with the majority discovered by the end of the late 1970s.
The enterobacterial common antigen (ECA) is a carbohydrate antigen found in the outer membrane of many Enterobacterales species. The antigen is unanimously absent from other gram-negative and gram-positive bacteria. Aeromonas hydrophila 209A is the only organism outside of Enterobacterales that expresses the ECA. More studies are needed to explain the presence of the antigen in this species as no other strains of this species express the antigen. The ECA is a polysaccharide made of repeating units of trisaccharides. The functions of these units have very few proven functions. Some evidence indicates role in pathogenicity in the bacteria that present the ECA. There are three separate types of ECA these include ECAPG, ECALPS, and ECACYC, each have different lengths. The synthesis of the ECA is controlled by the wec operon and has a 12-step synthesis which is described below. Due to the lack of proven function of the ECA, any clinical significance is hard to define however, some evidence suggests that human serum has antibodies against ECA.








