
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

Enzymes are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called enzymology and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties.
A diastase is any one of a group of enzymes that catalyses the breakdown of starch into maltose. For example, the diastase α-Amylase degrades starch to a mixture of the disaccharide maltose; the trisaccharide maltotriose, which contains three α (1-4)-linked glucose residues; and oligosaccharides, known as dextrins, that contain the α (1-6)-linked glucose branches.

In chemistry, a reagent or analytical reagent is a substance or compound added to a system to cause a chemical reaction, or test if one occurs. The terms reactant and reagent are often used interchangeably, but reactant specifies a substance consumed in the course of a chemical reaction. Solvents, though involved in the reaction mechanism, are usually not called reactants. Similarly, catalysts are not consumed by the reaction, so they are not reactants. In biochemistry, especially in connection with enzyme-catalyzed reactions, the reactants are commonly called substrates.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA topoisomerases are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one or both of the DNA strands. This transient break allows the DNA to be untangled or unwound, and, at the end of these processes, the DNA backbone is resealed. Since the overall chemical composition and connectivity of the DNA do not change, the DNA substrate and product are chemical isomers, differing only in their topology.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

DNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate".

Okazaki fragments are short sequences of DNA nucleotides which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.

Arthur Kornberg was an American biochemist who won the Nobel Prize in Physiology or Medicine 1959 for the discovery of "the mechanisms in the biological synthesis of ribonucleic acid and deoxyribonucleic acid" together with Spanish biochemist and physician Severo Ochoa of New York University. He was also awarded the Paul-Lewis Award in Enzyme Chemistry from the American Chemical Society in 1951, an L.H.D. degree from Yeshiva University in 1962, and the National Medal of Science in 1979. In 1991, Kornberg received the Golden Plate Award of the American Academy of Achievement and the Gairdner Foundation Award in 1995.

DNA polymerase II is a prokaryotic DNA-dependent DNA polymerase encoded by the PolB gene.
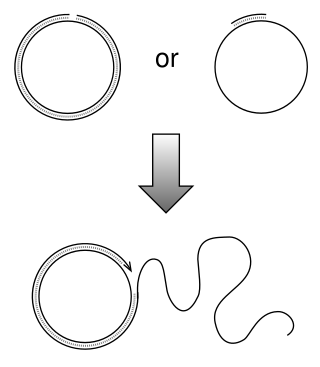
Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.

The history of biochemistry can be said to have started with the ancient Greeks who were interested in the composition and processes of life, although biochemistry as a specific scientific discipline has its beginning around the early 19th century. Some argued that the beginning of biochemistry may have been the discovery of the first enzyme, diastase, in 1833 by Anselme Payen, while others considered Eduard Buchner's first demonstration of a complex biochemical process alcoholic fermentation in cell-free extracts to be the birth of biochemistry. Some might also point to the influential work of Justus von Liebig from 1842, Animal chemistry, or, Organic chemistry in its applications to physiology and pathology, which presented a chemical theory of metabolism, or even earlier to the 18th century studies on fermentation and respiration by Antoine Lavoisier.

T7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.
The term proofreading is used in genetics to refer to the error-correcting processes, first proposed by John Hopfield and Jacques Ninio, involved in DNA replication, immune system specificity, enzyme-substrate recognition among many other processes that require enhanced specificity. The proofreading mechanisms of Hopfield and Ninio are non-equilibrium active processes that consume ATP to enhance specificity of various biochemical reactions.
Lorena Beese is a James B. Duke Professor of Biochemistry and Duke Cancer Institute Member. Her research involves structural mechanisms underlying DNA replication and repair, neurodegenerative diseases, cancer, and microbial pathogenesis; X-ray crystallography and cryo-electron microscopy; structure-based drug design; protein-protein and protein-nucleic acid interactions, enzyme mechanisms, chemical biology, protein structure and function.

DNA base flipping, or nucleotide flipping, is a mechanism in which a single nucleotide base, or nucleobase, is rotated outside the nucleic acid double helix. This occurs when a nucleic acid-processing enzyme needs access to the base to perform work on it, such as its excision for replacement with another base during DNA repair. It was first observed in 1994 using X-ray crystallography in a methyltransferase enzyme catalyzing methylation of a cytosine base in DNA. Since then, it has been shown to be used by different enzymes in many biological processes such as DNA methylation, various DNA repair mechanisms, and DNA replication. It can also occur in RNA double helices or in the DNA:RNA intermediates formed during RNA transcription.













