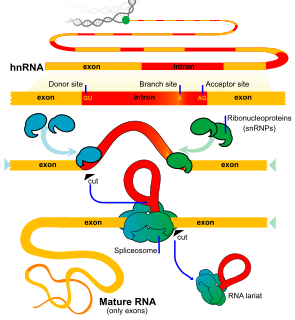
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2961 genes encoding RBPs in humans. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.
Post-transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms.

In molecular biology, SNORA2 is a non-coding RNA (ncRNA) which modifies other small nuclear RNAs (snRNAs). It is a member of the H/ACA class of small nucleolar RNA that guide the sites of modification of uridines to pseudouridines.

HIKESHI is a protein important in lung and multicellular organismal development that, in humans, is encoded by the HIKESHI gene. HIKESHI is found on chromosome 11 in humans and chromosome 7 in mice. Similar sequences (orthologs) are found in most animal and fungal species. The mouse homolog, lethal gene on chromosome 7 Rinchik 6 protein is encoded by the l7Rn6 gene.
Alternative Splicing Annotation Project (ASAP) in computational biology was a database for alternative splicing data maintained by the University of California from 2003 to 2013. The purpose of ASAP was to provide a source for data mining projects by consolidating the information generated by genomics and proteomics researchers.
The Alternative Splicing and Transcript Diversity database (ASTD) was a database of transcript variants maintained by the European Bioinformatics Institute from 2008 to 2012. It contained transcription initiation, polyadenylation and splicing variant data.
ECgene in computational biology is a database of genomic annotations taking alternative splicing events into consideration.
EDAS was a database of alternatively spliced human genes. It doesn't seem to exist anymore.
Hollywood is a RNA splicing database containing data for the splicing of orthologous genes in different species.
The Intronerator is a database of alternatively spliced genes and a database of introns for Caenorhabditis elegans.
SpliceInfo is a database for the four major alternative-splicing modes in the human genome.
De novo transcriptome assembly is the de novo sequence assembly method of creating a transcriptome without the aid of a reference genome.
Chimeric RNA, sometimes referred to as a fusion transcript, is composed of exons from two or more different genes that have the potential to encode novel proteins. These mRNAs are different from those produced by conventional splicing as they are produced by two or more gene loci.
WormBase is an online biological database about the biology and genome of the nematode model organism Caenorhabditis elegans and contains information about other related nematodes. WormBase is used by the C. elegans research community both as an information resource and as a place to publish and distribute their results. The database is regularly updated with new versions being released every two months. WormBase is one of the organizations participating in the Generic Model Organism Database (GMOD) project.

FAM163A, also known as cebelin and neuroblastoma-derived secretory protein (NDSP) is a protein that in humans is encoded by the FAM163A gene. This protein has been implicated in promoting proliferation and anchorage-independent growth of neuroblastoma cancer cells. In addition, this protein has been found to be up-regulated in the lung tissue of chronic smokers. FAM163A is found on human chromosome 1q25.2; its protein product is 167 amino acids long. FAM163A contains a very highly conserved signal peptide sequence, coded for by the first ~37 amino acids in its sequence; albeit only conserved in eukaryotes, the most distant of which being the Japanese Rice Fish.
In molecular phylogenetics, relationships among individuals are determined using character traits, such as DNA, RNA or protein, which may be obtained using a variety of sequencing technologies. High-throughput next-generation sequencing has become a popular technique in transcriptomics, which represent a snapshot of gene expression. In eukaryotes, making phylogenetic inferences using RNA is complicated by alternative splicing, which produces multiple transcripts from a single gene. As such, a variety of approaches may be used to improve phylogenetic inference using transcriptomic data obtained from RNA-Seq and processed using computational phylogenetics.

Alberto Kornblihtt is an Argentine molecular biologist who specializes in alternative ribonucleic acids splicing. Kornblihtt is credited with being among the first to document how a single transcribed gene can generate multiple protein variants. Kornblihtt was elected as a Foreign Associate of the National Academy of Sciences of the United States in 2011 and received the Diamond Award for the most relevant scientist of Argentina of the decade, alongside physicist Juan Martin Maldacena, in 2013.





