Related Research Articles

Methionine is an essential amino acid in humans.

Pyrrolysine is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group, a carboxylic acid group. Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH.
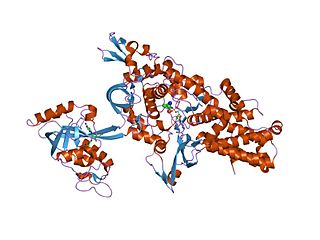
An aminoacyl-tRNA synthetase, also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code.
Succinyl-coenzyme A, abbreviated as succinyl-CoA or SucCoA, is a thioester of succinic acid and coenzyme A.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

In the mitochondrion, the matrix is the space within the inner membrane. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm. The mitochondrial matrix contains the mitochondrial DNA, ribosomes, soluble enzymes, small organic molecules, nucleotide cofactors, and inorganic ions.[1] The enzymes in the matrix facilitate reactions responsible for the production of ATP, such as the citric acid cycle, oxidative phosphorylation, oxidation of pyruvate, and the beta oxidation of fatty acids.

Succinyl coenzyme A synthetase is an enzyme that catalyzes the reversible reaction of succinyl-CoA to succinate. The enzyme facilitates the coupling of this reaction to the formation of a nucleoside triphosphate molecule from an inorganic phosphate molecule and a nucleoside diphosphate molecule. It plays a key role as one of the catalysts involved in the citric acid cycle, a central pathway in cellular metabolism, and it is located within the mitochondrial matrix of a cell.

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).
In enzymology, a tryptophan-tRNA ligase is an enzyme that catalyzes the chemical reaction

In enzymology, a cystathionine gamma-synthase is an enzyme that catalyzes the formation of cystathionine from cysteine and an activated derivative of homoserine, e.g.:
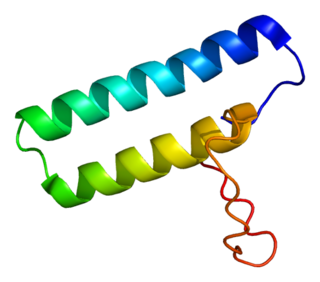
Histidyl-tRNA synthetase (HARS) also known as histidine-tRNA ligase, is an enzyme which in humans is encoded by the HARS gene.

Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial (SUCLA2), also known as ADP-forming succinyl-CoA synthetase (SCS-A), is an enzyme that in humans is encoded by the SUCLA2 gene on chromosome 13.

Probable histidyl-tRNA synthetase, mitochondrial is an enzyme that in humans is encoded by the HARS2 gene.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.

The sucC RNA motif is a conserved RNA structure discovered using bioinformatics. sucC RNAs are found in the genus Pseudomonas, and are consistently found in possible 5' untranslated regions of sucC genes. These genes encode Succinyl coenzyme A synthetase, and are hypothesised to be regulated by the sucC RNAs. sucC genes participate in the citric acid cycle, and another gene involved in the citric acid cycle, sucA, is also predicted to be regulated by a conserved RNA structure.
In molecular biology, Toxic Small RNA(tsRNA, not to be confused with tRNA-derived small RNA) is a family of trans-encoded small non-coding RNA found exclusively in intergenic regions of Betaproteobacteria. Several paralogous loci may encode similar tsRNAs in each coding genome. Typically, each species of Burkholderia has 3-5 homologous tsRNAs. Experiments with four species of the Burkholderia lineage showed conserved and constitutive expression of tsRNAs in logarithmic growth phases.
Streptomyces albidoflavus is a bacterium species from the genus of Streptomyces which has been isolated from soil from Poland. Streptomyces albidoflavus produces dibutyl phthalate and streptothricins.

Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial is an enzyme that in humans is encoded by the SUCLG2 gene on chromosome 3.
NmsRA and NmsRB, RcoF1 and RcoF2 as well as NgncR_162 and NgncR_163 are all names of neisserial sibling small regulatory RNAs described and independently named in three publications. NmsRB/RcoF1/NgncR_163 was shown to be the predominant sibling. The sRNAs are tandemly arranged, structurally nearly identical and share 70% sequence identity. They translationally down-regulate genes involved in basic metabolic processes including tricarboxylic acid cycle enzymes and amino acid uptake and degradation. The target genes include: fumC, sdhC, gltA, sucC, prpB and prpC. The expression of the sRNAs is presumably under the control of RelA, as shown for N. meningitidis. Furthermore, the sRNAs interact with Hfq protein and target repression of putative colonization factor of the human nasopharynx PrpB mRNA, hence one of the proposed names is RNA regulating colonization factor.

The Rothia-sucC RNA motif is a conserved RNA structure that was discovered by bioinformatics. Rothia-sucC motif RNAs are found in the actinobacterial genus Rothia.
References
- ↑ Schilling, D; Findeiss, S; Richter, AS; Taylor, JA; Gerischer, U (September 2010). "The small RNA Aar in Acinetobacter baylyi: a putative regulator of amino acid metabolism". Archives of Microbiology. 192 (9): 691–702. Bibcode:2010ArMic.192..691S. doi:10.1007/s00203-010-0592-6. PMID 20559624. S2CID 16709707.
- ↑ Schimmel, PR; Söll, D (1979). "Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs". Annual Review of Biochemistry. 48: 601–648. doi:10.1146/annurev.bi.48.070179.003125. PMID 382994.
- ↑ Buck, D; Guest, JR (June 15, 1969). "Overexpression and site-directed mutagenesis of the succinyl-CoA synthetase of Escherichia coli and nucleotide sequence of a gene (g30) that is adjacent to the suc operon". The Biochemical Journal. 260 (3): 737–747. doi:10.1042/bj2600737. PMC 1138739 . PMID 2548486.
- ↑ Schilling, D; Findeiss, S; Richter, AS; Taylor, JA; Gerischer, U (September 2010). "The small RNA Aar in Acinetobacter baylyi: a putative regulator of amino acid metabolism". Archives of Microbiology. 192 (9): 691–702. Bibcode:2010ArMic.192..691S. doi:10.1007/s00203-010-0592-6. PMID 20559624. S2CID 16709707.