In molecular biology, small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing.

In molecular biology, Small nucleolar RNA snoR639 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a 'guide RNA'.

In molecular biology, SNORA66 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA". U66 was originally cloned from HeLa cells and belongs to the H/ACA box class of snoRNAs as it has the predicted hairpin-hinge-hairpin-tail structure, has the conserved H/ACA-box motifs and is found associated with GAR1 protein. U66 is predicted to guide the pseudouridylation of U119 of 18S ribosomal RNA (rRNA). Pseudouridylation is the to the different isomeric form pseudouridine.

In molecular biology, Small nucleolar RNA SNORA68 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

In molecular biology, Small nucleolar RNA SNORA69 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

In molecular biology, small nucleolar RNA SNORA72 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

In molecular biology, Small nucleolar RNA SNORA70 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

The Actino-pnp RNA motif is a conserved structure found in Actinomycetota that is apparently in the 5' untranslated regions of genes predicted to encode exoribonucleases. The RNA element's function is likely analogous to an RNA structure found upstream of polynucleotide phosphorylase genes in E. coli and related enterobacteria. In this latter system, the polynucleotide phosphorlyase gene regulates its own expression levels by a feedback mechanism that involves its activity upon the RNA structure. However, the E. coli RNA appears to be structurally unrelated to the Actino-pnp motif.
DUF3800 RNA motifs refer to conserved RNA structures that were discovered by bioinformatics and are usually located nearby to genes that encode proteins containing the conserved domain called "DUF3800". However, the gene can be located 5′ or 3′ relative to the RNA, and could be on the same or on the opposite DNA strand. This arrangement is typical of RNA and proteins that function as an RNA-protein complex, but could have other explanations. The function of this protein domain is unknown, and no specific biological function has been proposed for the RNA. However, after the detection of the RNA motifs, DUF3800 domains were predicted to be involved with retrons.
The DABA-DC-AT RNA motif is a conserved RNA structure that was discovered by bioinformatics. DABA-DC-AT motifs are found in Gammaproteobacteria, but overwhelmingly in the order Vibrionales.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The Fibrobacter-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Fibrobacter-1 motifs are found in organisms in the genus Fibrobacter, and in many metagenomic sequences.
The freshwater-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Freshwater-2 motif RNAs are found in metagenomic sequences that are isolated from aquatic and especially freshwater environments. As of 2018, no freshwater-2 RNA has been identified in a classified organism.
The Fusobacteriales-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Fusobacteriales-1 motif RNAs are found in Fusobacteriales.

The IMPDH RNA motif is a conserved RNA structure that was discovered by bioinformatics. IMPDH motif RNAs are found in organisms classified within the genus Faecalibacterium.
lysM RNA motifs are conserved RNA structures that were discovered by bioinformatics. Such bacterial motifs are defined by consistently being upstream of 'lysM' genes, which encode lysin protein domains, a conserved domain that participates in cell wall degradation. lysM motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes, although this hypothesis is not certain.

The MDR-NUDIX RNA motif is a conserved RNA structure that was discovered by bioinformatics. The MDR-NUDIX motif is found in the poorly studied phylum TM7.
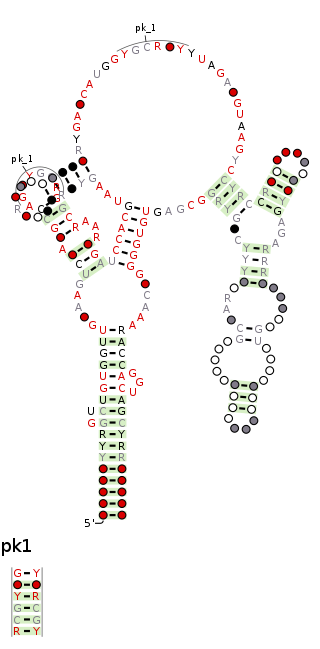
The raiA RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA motif RNAs are found in Actinomycetota and Bacillota, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the raiA RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise.
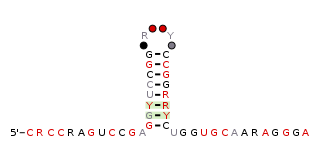
The raiA-hairpin RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA-hairpin motif RNAs are found in organism classified within the genus Streptomyces.

The ssNA-helicase RNA motif is a conserved RNA structure that was discovered by bioinformatics. Although the ssNA-helicase motif was published as an RNA candidate, there is some reason to suspect that it might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available.














