
Magnetic resonance imaging (MRI) is a medical imaging technique used in radiology to form pictures of the anatomy and the physiological processes of the body. MRI scanners use strong magnetic fields, magnetic field gradients, and radio waves to generate images of the organs in the body. MRI does not involve X-rays or the use of ionizing radiation, which distinguishes it from computed tomography (CT) and positron emission tomography (PET) scans. MRI is a medical application of nuclear magnetic resonance (NMR) which can also be used for imaging in other NMR applications, such as NMR spectroscopy.

Functional magnetic resonance imaging or functional MRI (fMRI) measures brain activity by detecting changes associated with blood flow. This technique relies on the fact that cerebral blood flow and neuronal activation are coupled. When an area of the brain is in use, blood flow to that region also increases.
Functional integration is the study of how brain regions work together to process information and effect responses. Though functional integration frequently relies on anatomic knowledge of the connections between brain areas, the emphasis is on how large clusters of neurons – numbering in the thousands or millions – fire together under various stimuli. The large datasets required for such a whole-scale picture of brain function have motivated the development of several novel and general methods for the statistical analysis of interdependence, such as dynamic causal modelling and statistical linear parametric mapping. These datasets are typically gathered in human subjects by non-invasive methods such as EEG/MEG, fMRI, or PET. The results can be of clinical value by helping to identify the regions responsible for psychiatric disorders, as well as to assess how different activities or lifestyles affect the functioning of the brain.

Diffusion-weighted magnetic resonance imaging is the use of specific MRI sequences as well as software that generates images from the resulting data that uses the diffusion of water molecules to generate contrast in MR images. It allows the mapping of the diffusion process of molecules, mainly water, in biological tissues, in vivo and non-invasively. Molecular diffusion in tissues is not random, but reflects interactions with many obstacles, such as macromolecules, fibers, and membranes. Water molecule diffusion patterns can therefore reveal microscopic details about tissue architecture, either normal or in a diseased state. A special kind of DWI, diffusion tensor imaging (DTI), has been used extensively to map white matter tractography in the brain.

Magnetic resonance angiography (MRA) is a group of techniques based on magnetic resonance imaging (MRI) to image blood vessels. Magnetic resonance angiography is used to generate images of arteries in order to evaluate them for stenosis, occlusions, aneurysms or other abnormalities. MRA is often used to evaluate the arteries of the neck and brain, the thoracic and abdominal aorta, the renal arteries, and the legs.

Magnetic resonance microscopy is magnetic resonance imaging (MRI) at a microscopic level down to the scale of microns. The first definition of MRM was MRI having voxel resolutions of better than 100 μm.
Fast low angle shot magnetic resonance imaging is a particular sequence of magnetic resonance imaging. It is a gradient echo sequence which combines a low-flip angle radio-frequency excitation of the nuclear magnetic resonance signal with a short repetition time. It is the generic form of steady-state free precession imaging.
In vivo magnetic resonance spectroscopy (MRS) is a specialized technique associated with magnetic resonance imaging (MRI).

Real-time magnetic resonance imaging (RT-MRI) refers to the continuous monitoring ("filming") of moving objects in real time. Because MRI is based on time-consuming scanning of k-space, real-time MRI was possible only with low image quality or low temporal resolution. Using an iterative reconstruction algorithm these limitations have recently been removed: a new method for real-time MRI achieves a temporal resolution of 20 to 30 milliseconds for images with an in-plane resolution of 1.5 to 2.0 mm. Real-time MRI promises to add important information about diseases of the joints and the heart. In many cases MRI examinations may become easier and more comfortable for patients.

Quantitative susceptibility mapping (QSM) provides a novel contrast mechanism in magnetic resonance imaging (MRI) different from traditional susceptibility weighted imaging. The voxel intensity in QSM is linearly proportional to the underlying tissue apparent magnetic susceptibility, which is useful for chemical identification and quantification of specific biomarkers including iron, calcium, gadolinium, and super paramagnetic iron oxide (SPIO) nano-particles. QSM utilizes phase images, solves the magnetic field to susceptibility source inverse problem, and generates a three-dimensional susceptibility distribution. Due to its quantitative nature and sensitivity to certain kinds of material, potential QSM applications include standardized quantitative stratification of cerebral microbleeds and neurodegenerative disease, accurate gadolinium quantification in contrast enhanced MRI, and direct monitoring of targeted theranostic drug biodistribution in nanomedicine.

Magnetic resonance imaging of the brain uses magnetic resonance imaging (MRI) to produce high quality two-dimensional or three-dimensional images of the brain and brainstem as well as the cerebellum without the use of ionizing radiation (X-rays) or radioactive tracers.
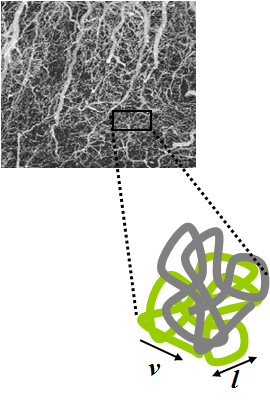
Intravoxel incoherent motion (IVIM) imaging is a concept and a method initially introduced and developed by Le Bihan et al. to quantitatively assess all the microscopic translational motions that could contribute to the signal acquired with diffusion MRI. In this model, biological tissue contains two distinct environments: molecular diffusion of water in the tissue, and microcirculation of blood in the capillary network (perfusion). The concept introduced by D. Le Bihan is that water flowing in capillaries mimics a random walk (Fig.1), as long as the assumption that all directions are represented in the capillaries is satisfied.
Medical image computing (MIC) is an interdisciplinary field at the intersection of computer science, information engineering, electrical engineering, physics, mathematics and medicine. This field develops computational and mathematical methods for solving problems pertaining to medical images and their use for biomedical research and clinical care.
Functional magnetic resonance spectroscopy of the brain (fMRS) uses magnetic resonance imaging (MRI) to study brain metabolism during brain activation. The data generated by fMRS usually shows spectra of resonances, instead of a brain image, as with MRI. The area under peaks in the spectrum represents relative concentrations of metabolites.
Synthetic MRI is a simulation method in Magnetic Resonance Imaging (MRI), for generating contrast weighted images based on measurement of tissue properties. The synthetic (simulated) images are generated after an MR study, from parametric maps of tissue properties. It is thereby possible to generate several contrast weightings from the same acquisition. This is different from conventional MRI, where the signal acquired from the tissue is used to generate an image directly, often generating only one contrast weighting per acquisition. The synthetic images are similar in appearance to those normally acquired with an MRI scanner.
The history of magnetic resonance imaging (MRI) includes the work of many researchers who contributed to the discovery of nuclear magnetic resonance (NMR) and described the underlying physics of magnetic resonance imaging, starting early in the twentieth century. One researcher was American physicist Isidor Isaac Rabi who won the Nobel Prize in Physics in 1944 for his discovery of nuclear magnetic resonance, which is used in magnetic resonance imaging. MR imaging was invented by Paul C. Lauterbur who developed a mechanism to encode spatial information into an NMR signal using magnetic field gradients in September 1971; he published the theory behind it in March 1973.

An MRI sequence in magnetic resonance imaging (MRI) is a particular setting of pulse sequences and pulsed field gradients, resulting in a particular image appearance.
An MRI artifact is a visual artifact in magnetic resonance imaging (MRI). It is a feature appearing in an image that is not present in the original object. Many different artifacts can occur during MRI, some affecting the diagnostic quality, while others may be confused with pathology. Artifacts can be classified as patient-related, signal processing-dependent and hardware (machine)-related.
Arterial spin labeling (ASL), also known as arterial spin tagging, is a magnetic resonance imaging technique used to quantify cerebral blood perfusion by labelling blood water as it flows throughout the brain. ASL specifically refers to magnetic labeling of arterial blood below or in the imaging slab, without the need of gadolinium contrast. A number of ASL schemes are possible, the simplest being flow alternating inversion recovery (FAIR) which requires two acquisitions of identical parameters with the exception of the out-of-slice saturation; the difference in the two images is theoretically only from inflowing spins, and may be considered a 'perfusion map'. The ASL technique was developed by Alan P. Koretsky, Donald S. Williams, John A. Detre and John S. Leigh Jr in 1992.
Magnetic resonance fingerprinting (MRF) is methodology in quantitative magnetic resonance imaging (MRI) characterized by a pseudo-randomized acquisition strategy. It involves creating unique signal patterns or 'fingerprints' for different materials or tissues after which a pattern recognition algorithm matches these fingerprints with a predefined dictionary of expected signal patterns. This process translates the data into quantitative maps, revealing information about the magnetic properties being investigated.









