| Antizyme RNA frameshifting stimulation element | |
|---|---|
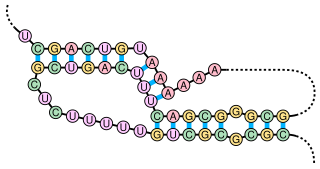
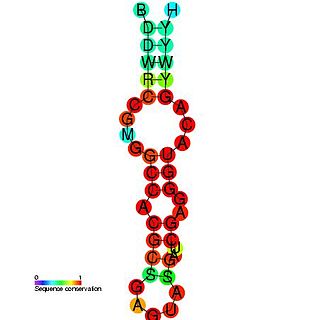
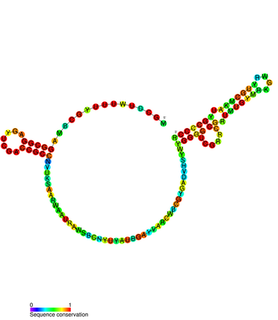
 Predicted secondary structure and sequence conservation of Antizyme_FSE | |
| Identifiers | |
| Symbol | Antizyme_FSE |
| Rfam | RF00381 |
| Other data | |
| RNA type | Cis-reg; frameshift_element |
| Domain(s) | Eukaryota |
| SO | 0000233 |
| PDB structures | PDBe |
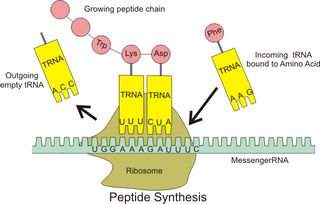
Antizyme RNA frameshifting stimulation element is a structural element which is found in antizyme mRNA and is known to promote frameshifting. Antizyme genes have two partially overlapping open reading frames, the second, which encodes the functional (antizyme) protein requires +1 translational frameshifting. This frameshift is stimulated by a pseudoknot present 3' of the frameshift site in the antizyme mRNA. The frameshifting efficiency is dependent on the concentration of polyamines in the cell, when the polyamine concentration is high frameshifting is more likely to occur which leads to an increase in the quantity of functional antizyme produced. The functional antizyme acts to reduce ornithine decarboxylase (ODC) activity which leads to a drop in polyamines present in the cell. Therefore, this family can be thought of as a biosensor for intracellular free polyamines that functions via a negative feedback loop. [1]
Cis-regulatory elements (CREs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.

In molecular biology, Ornithine decarboxylase antizyme (ODC-AZ) is an ornithine decarboxylase inhibitor. It binds to, and destabilises, ornithine decarboxylase (ODC), a key enzyme in polyamine synthesis. ODC is then rapidly degraded. It was first characterized in 1981. The expression of ODC-AZ requires programmed, ribosomal frameshifting which is modulated according to the cellular concentration of polyamines. High levels of polyamines induce a +1 ribosomal frameshift in the translation of mRNA for the antizyme leading to the expression of a full-length protein. At least two forms of ODC-AZ exist in mammals and the protein has been found in Drosophila as well as in Saccharomyces yeast.
In molecular genetics, an open reading frame (ORF) is the part of a reading frame that has the ability to be translated. An ORF is a continuous stretch of codons that begins with a start codon and ends at a stop codon. An ATG codon within the ORF may indicate where translation starts. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final mRNA for protein translation. In the context of gene finding, the start-stop definition of an ORF therefore only applies to spliced mRNAs, not genomic DNA, since introns may contain stop codons and/or cause shifts between reading frames. An alternative definition says that an ORF is a sequence that has a length divisible by three and is bounded by stop codons. This more general definition can also be useful in the context of transcriptomics and/or metagenomics, where start and/or stop codon may not be present in the obtained sequences. Such an ORF corresponds to parts of a gene rather than the complete gene.