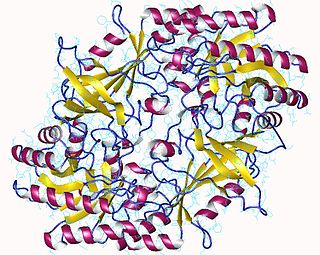
The enzyme ornithine decarboxylase catalyzes the decarboxylation of ornithine to form putrescine. This reaction is the committed step in polyamine synthesis. In humans, this protein has 461 amino acids and forms a homodimer.
In molecular genetics, the Krüppel-like family of transcription factors (KLFs) are a set of eukaryotic C2H2 zinc finger DNA-binding proteins that regulate gene expression. This family has been expanded to also include the Sp transcription factor and related proteins, forming the Sp/KLF family.

Antizyme RNA frameshifting stimulation element is a structural element which is found in antizyme mRNA and is known to promote frameshifting. Antizyme genes have two partially overlapping open reading frames, the second, which encodes the functional (antizyme) protein requires +1 translational frameshifting. This frameshift is stimulated by a pseudoknot present 3' of the frameshift site in the antizyme mRNA. The frameshifting efficiency is dependent on the concentration of polyamines in the cell, when the polyamine concentration is high frameshifting is more likely to occur which leads to an increase in the quantity of functional antizyme produced. The functional antizyme acts to reduce ornithine decarboxylase (ODC) activity which leads to a drop in polyamines present in the cell. Therefore, this family can be thought of as a biosensor for intracellular free polyamines that functions via a negative feedback loop.

In molecular biology, the coronavirus frameshifting stimulation element is a conserved stem-loop of RNA found in coronaviruses that can promote ribosomal frameshifting. Such RNA molecules interact with a downstream region to form a pseudoknot structure; the region varies according to the virus but pseudoknot formation is known to stimulate frameshifting. In the classical situation, a sequence 32 nucleotides downstream of the stem is complementary to part of the loop. In other coronaviruses, however, another stem-loop structure around 150 nucleotides downstream can interact with members of this family to form kissing stem-loops and stimulate frameshifting.

The DnaX ribosomal frameshifting element is a RNA element found in the mRNA of the dnaX gene in E. coli. The dnaX gene has two encoded products, tau and gamma, which are produced in a 1:1 ratio. The gamma protein is synthesised due to programmed frameshifting and is shorter than tau. The two products of the dnaX gene are DNA polymerase III subunits.
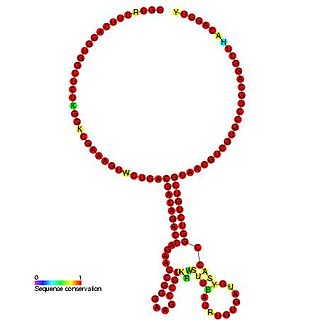
The Insertion sequence IS1222 ribosomal frameshifting element is an RNA element found in the insertion sequence IS222. The ribosomal frameshifting element stimulates frameshifting which is known to be required for transposition.
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.

40S ribosomal protein S3a is a protein that in humans is encoded by the RPS3A gene.

S-methyl-5'-thioadenosine phosphorylase (MTAP) is an enzyme in humans responsible for polyamine metabolism. It is encoded by the methylthioadenosine phosphorylase (MTAP) gene on chromosome 9. Multiple alternatively spliced transcript variants have been described for this gene, but their full-length natures remain unknown.

Ornithine decarboxylase antizyme is an enzyme that in humans is encoded by the OAZ1 gene.

The RPS2 gene is the gene which, in humans, encodes the 40S ribosomal protein S2.

40S ribosomal protein S25 (eS25) is a protein that in humans is encoded by the RPS25 gene.

60S ribosomal protein L22 is a protein that in humans is encoded by the RPL22 gene on Chromosome 1.

Protein odd-skipped-related 1 is a transcription factor that in humans is encoded by the OSR1 gene. The OSR1 and OSR2 transcription factors participate in the normal development of body parts such as the kidney.

Antizyme inhibitor 2 (AzI2) also erroneously known as arginine decarboxylase (ADC) is a protein that in humans is encoded by the AZIN2 gene. In contrast to initial suggestions, Antizyme inhibitor 2 does not act as arginine decarboxylase (ADC) in mammalian cells

Antizyme inhibitor 1 is a protein that in humans is encoded by the AZIN1 gene.

Ornithine decarboxylase antizyme 2 is an enzyme that in humans is encoded by the OAZ2 gene.
A polyamine is an organic compound having more than two amino groups. Alkyl polyamines occur naturally, but some are synthetic. Alkylpolyamines are colorless, hygroscopic, and water soluble. Near neutral pH, they exist as the ammonium derivatives. Most aromatic polyamines are crystalline solids at room temperature.
John Atkins is a research professor at University College Cork and a member of the Royal Irish Academy since 2003. Atkins was the first Irish national to be elected as a member of the EMBO Organization. In 2002 Science Foundation Ireland appointed Atkins as its first Director of Biotechnology. Atkins is also an honorary Professor of Genetics at his alma mater Trinity College, Dublin.
In molecular biology, the ODC internal ribosome entry site (IRES) is an RNA element present in the 5′ UTR of the mRNA encoding ornithine decarboxylase. It has been suggested that this IRES allows cap-independent translation of ornithine decarboxylase at the G2/M phase of the cell cycle, however there is some doubt about this. Translation from this IRES is activated by the zinc finger protein ZNF9 and by Poly(rC)-binding protein 2 (PCBP2). It is also activated in Ras-transformed cells.














